Metagenomic Discovery and the Size of DNA Space
How much of the DNA in the world has been sequenced? How much biodiversity has been explored? If you're a developer trying to build things with biology, natural DNA sequences are an incredible resource, but also a mysterious and largely unexplored one. Today we're talking about metagenomic discovery and what it takes to use DNA databases effectively.
Transcript
Let's say you need a better enzyme for a biotech application. Generally speaking, there are two big strategies for getting it. You can use protein engineering to modify an existing enzyme. Or you can find a new enzyme out there in nature, a process called metagenomic discovery.
Here at Ginkgo we do both, often at the same time. That's the beauty of the foundry, we have the capacity to explore different strategies when we need to. But the metagenomic discovery problem in particular is interesting to me as a kind of thought experiment in natural philosophy.
It's like we're looking at all the enzymes that have ever been described by science and saying "yeah but maybe not." We're betting that something out there, in all of nature, can be better than anything we know. But what does "out there" look like? What are the odds that something out there is better? How can we improve those odds?
I think it's easy to overestimate how thoroughly we've explored DNA sequence space. I'm guilty of it. How many times in my career have I gone looking for an enzyme, taken it totally for granted that it had already been described, and just kind of assumed that one would be good enough? The existing databases are big enough to give the impression they've got it all. In other words, BLAST spoiled us.
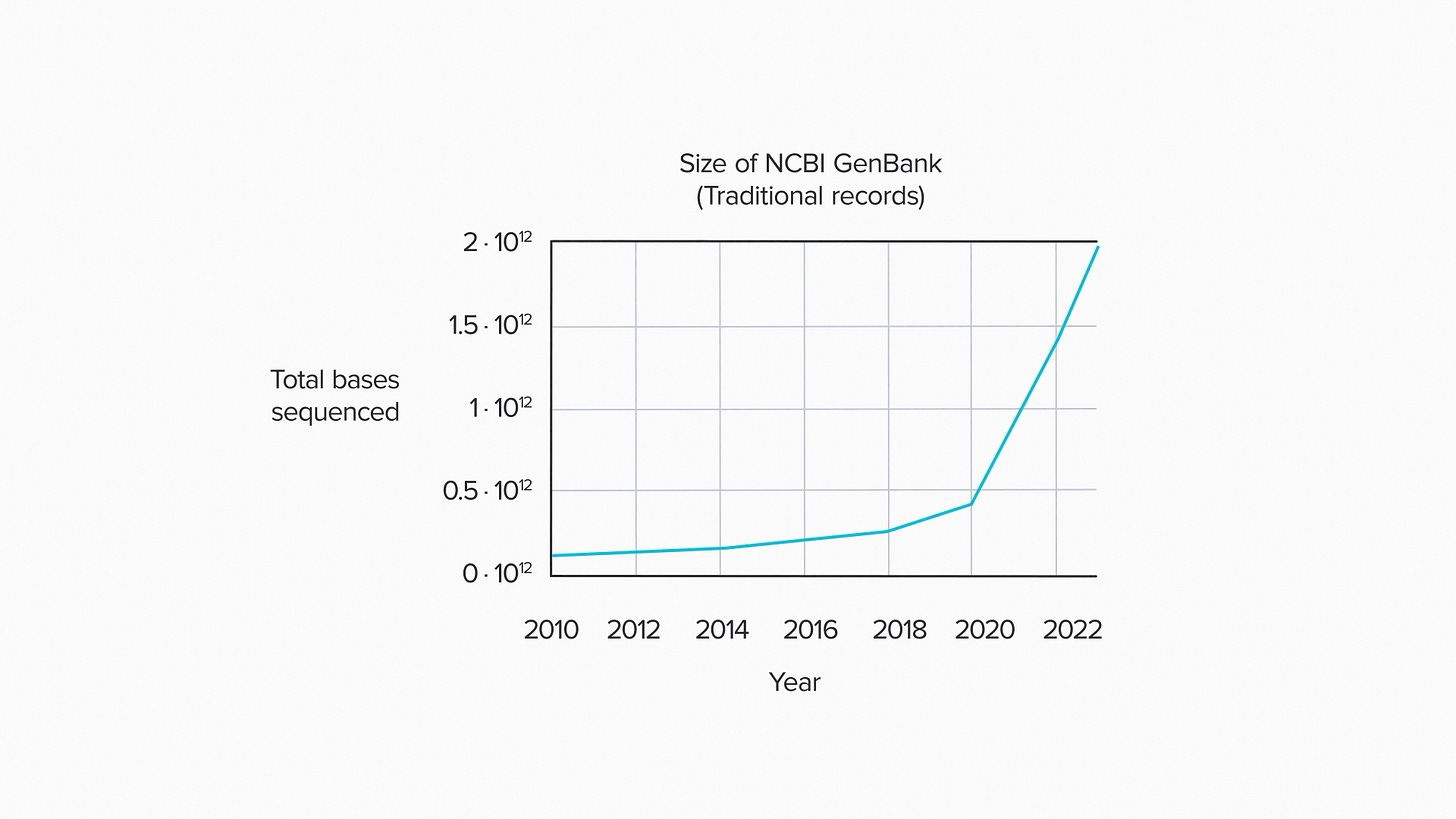
Take a look at GenBank for example, the largest public DNA sequence database. It's been growing exponentially since 1982. As of 2023, there were about 2·10¹² base pairs of sequence data in GenBank1. That's about 2 terabases of DNA - a million million nucleotides.
Which seems like a lot. But how does it compare to nature's DNA database? In 2015, a published estimate for the total amount of sequence information on earth was more like 10³⁷ base pairs2. Now I try not to get too technical in these videos, but according to my calculations, that's approximately infinity bajillion times larger than GenBank.
It's not just that nature is larger than what we know, it's that nature is so much larger we can't really conceptualize it. The implication of this for metagenomic discovery is that new DNA sequences are effectively a bottomless well. Whatever is the limit of new sequence information, we're not close to it. A particular type of enzyme might already be described, but nature is full of other variants with different sequences, different structures and different functional profiles.
That's the motivation here at Ginkgo to put together our own DNA database to support customer projects. We call it the UMDB - the Unified Metagenomics Database. Now, I'm about to share the kind of info that goes obsolete fast, because new data is coming in all the time. But as of this moment, we've got about 2 billion genes worth of proprietary sequence data to use for discovery. That compares to about 250 million genes in a well known public database like UniProt.
Here's a recent example of how metagenomic discovery played out in practice. A customer came to us because they wanted to make an enzyme. This was a dehydrogenase to be used as part of a chemical process.
The chemical reaction was specific to their application, but the overall enzyme requirements were pretty typical. They needed something with high activity, that was specific to their target molecule, and that would work under their process conditions. There were known examples of the enzyme in public sequence databases, but they were too slow. They needed something faster.
So we searched the public databases, combined with our own UMDB, and identified 500 candidate enzymes. We built the DNA to express them all and tested them for activity.
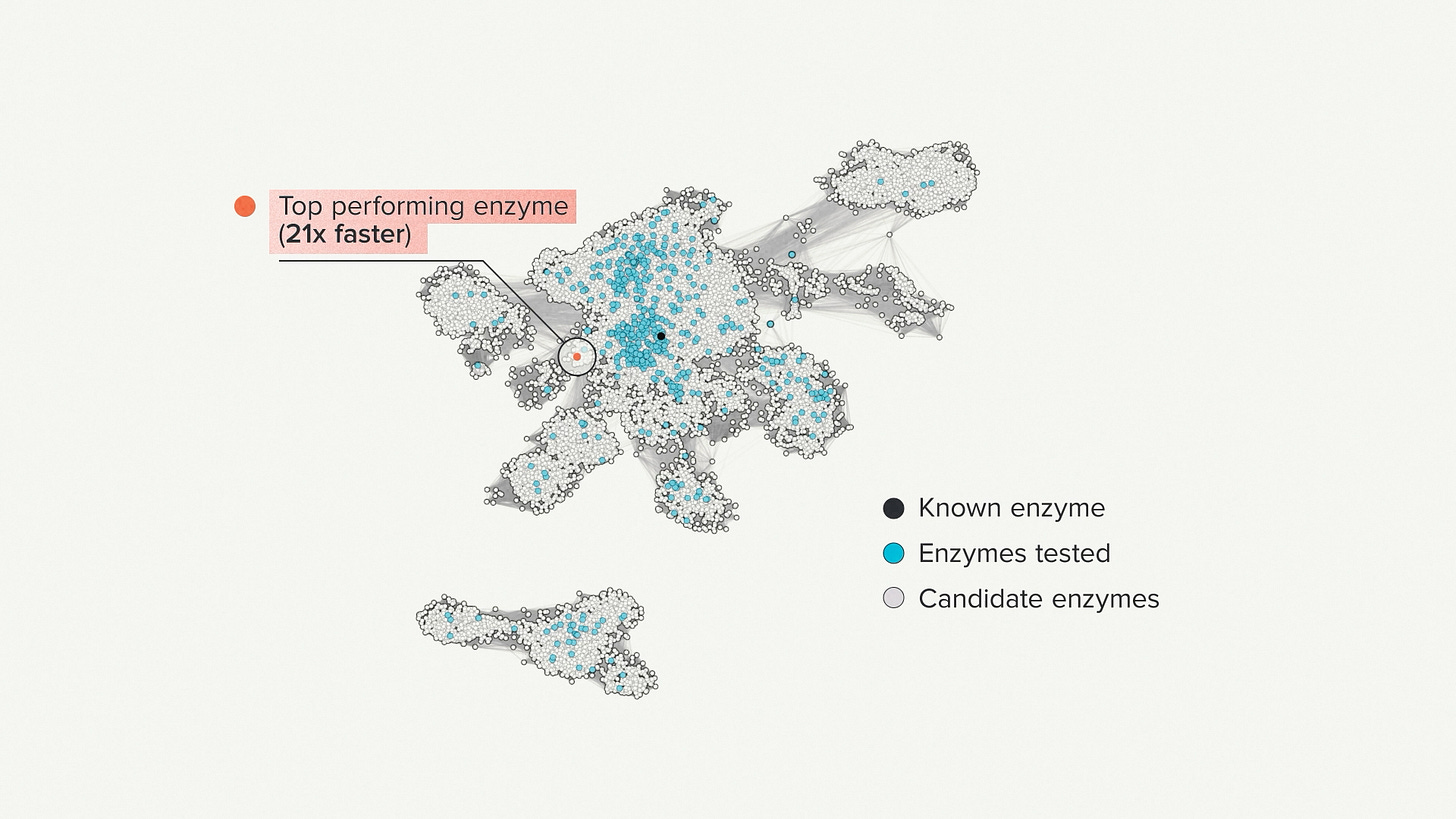
Here I'm showing a projection of enzyme sequence space. It's one way to visualize the vastness of nature, at least for the tiny little part that was relevant for this project. The black dot is an enzyme that was already known. We call that the seed sequence. The blue dots represent the enzymes that we selected to synthesize and test - 500 in total. The red dot is the top performer.
There was very little sequence similarity between the seed enzyme that we started with and the one we eventually found. In this case it was about 33% sequence match - a completely different part of the evolutionary tree. It performed the same reaction as the seed, only 21 times faster. This was a great result - just what the customer was looking for and better than their performance targets.
Now this was just one project. Metagenomic search doesn't always hit like this. But I think it illustrates some more general principles about what it takes to search DNA effectively. It comes down to:
The sheer size of the DNA library you can search. This is pretty straightforward. The great enzyme sequences have to be there before you can find them.
The quality of your AI-guided search tools. This is how you get from a very big enzyme universe to a manageable number of candidates for testing.
Your capacity for testing. Because search is always going to give you lots of possibilities but ultimately it takes experiments to know what is real.
For best results we want all of the above. More of everything.
Some day, I guess, we'll start to find the limits of what functions are discoverable in DNA. Our DNA databases will start to cover all or most of the natural diversity. Our AI tools will be able to separate the very good candidates from the very very good candidates. Our automation capacity will be able to test everything we're curious about.
But we're far away from that. We're still in the world where we've only explored a tiny fraction of what nature has to offer. As long as that is the case, metagenomic discovery is going to keep getting better with scale.