What happens when the unstoppable force of AI meets the immoveable object of biological hard tech?
Transcript
We're at a really interesting moment for AI in biotech where genuinely new capabilities are meeting real world applications and nobody knows how impactful they're going to be in practice. It's almost a case of the unstoppable force meets the immovable object: AI, very powerful; versus biological problems, very hard. Who wins?
I have my personal experience engineering biology and my own expectations about what's possible and what's realistic. And then I saw some results recently that overturned those expectations. Let me share the case study.
We were doing an enzyme engineering project for a pharma customer. We started from zero by discovering an enzyme in nature that could do a reaction never described before. We used a few different flavors of AI to improve its performance: making it more active and also more specific. For simplicity, I'm going to focus on the activity improvements.
The project played out in 7 iterations of about 8 weeks each. In each round, we generated a library of sequence variants of the enzyme and measured their activity. We measured performance for a few hundred or a few thousand enzymes per round. Those measurements get fed back into the AI model for the next generation.
The question is: how should we expect a project like this to go? How much improvement should we anticipate from each round? Biology will always surprise us and no two projects will be exactly the same. So we have to rely, before the data starts coming in, on our intuitions about what is possible.
Traditionally, my intuition would be modest improvements with diminishing returns. I'd expect a decent improvement in the first round, a moderate improvement in the second, and then a pretty rapid leveling off. I think most biologists would share this expectation because this shape of curve comes up everywhere in our training.
We called it saturation kinetics in biochemistry. Or a sigmoid curve in cell biology. If you're growing tomatoes, the first bit of fertilizer helps a lot. The second bit helps a little and pretty soon it's pointless to add more. It's the same pattern for nutrient uptake by a bacterium. Or neurotransmitters binding to receptors. Or the dose-response curve of a drug. It's a good default assumption when you change biology, whatever you do to change it will have progressively less impact.
Contrast this to the AI space where the scaling laws seem to keep going up. The more resources you can throw at a model, the better it performs. More compute, more parameters, more training data. We're used to seeing AI model inputs scale into the billions and beyond and continue to yield performance returns. Now - it has to stop somewhere. Nothing can grow forever. But in the case of AI, people are comfortable betting that, wherever that limit is, we're not close to it. Whereas in biology, people usually bet that the limit is relatively close by.
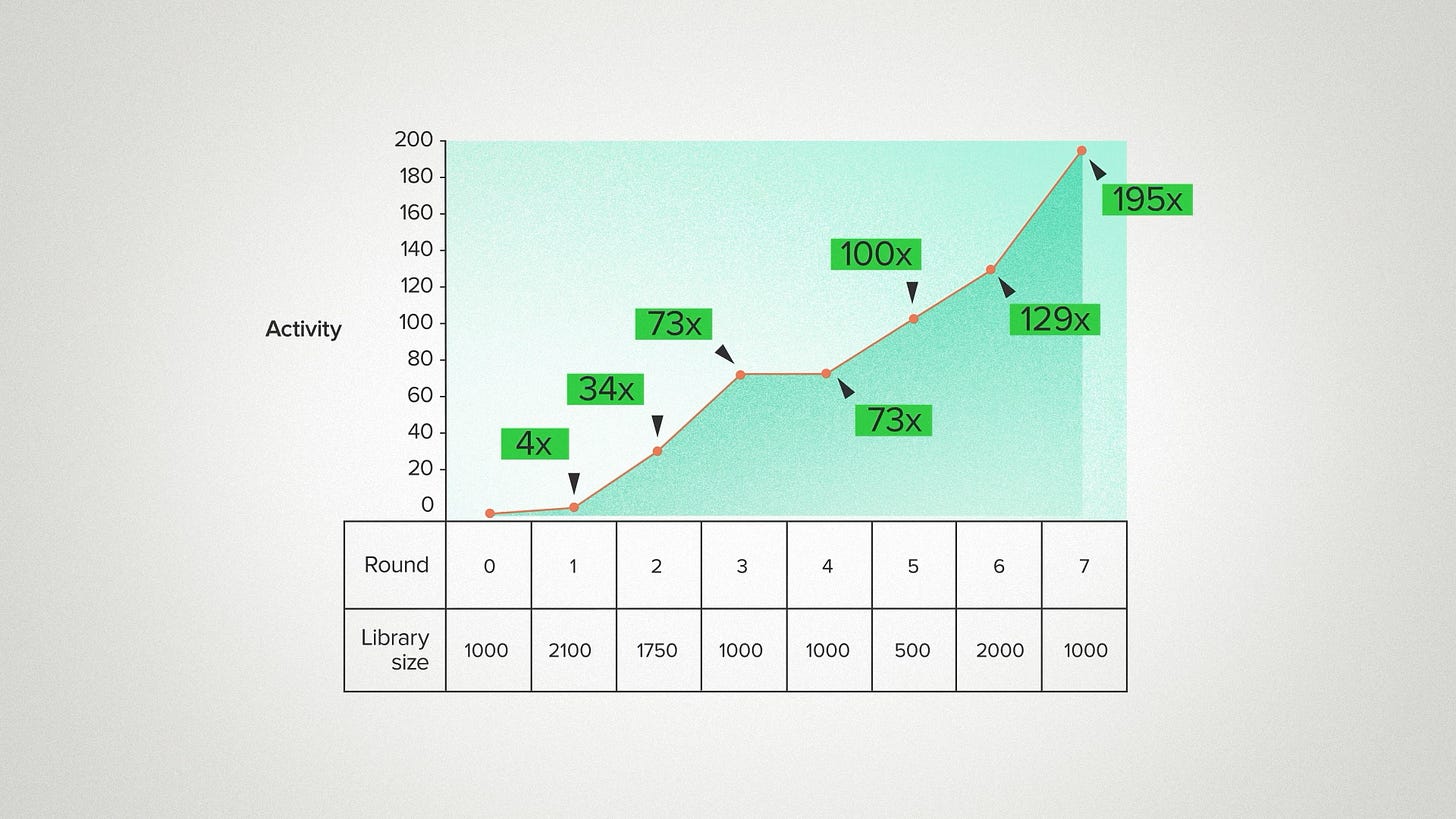
So that gives us two mental models. Diminishing returns vs. endless scaling. Which one wins? At least for this particular enzyme? Did the performance keep growing with the size of the dataset, like an AI developer might expect? Or did it level off after 2-3 design cycles, like a biology developer might expect?
It looked like this: 4x improvement in the first round. 34x in the second. Then 73 for the next two rounds - it seems like it might be done. But then 100, 129, 195. Can it go on like this forever? No - it can't. But it really is remarkable to me how much this enzyme was improved and for how long it kept getting better. Back in my day, none of my synthetic biology projects went past 2 or 3 rounds.
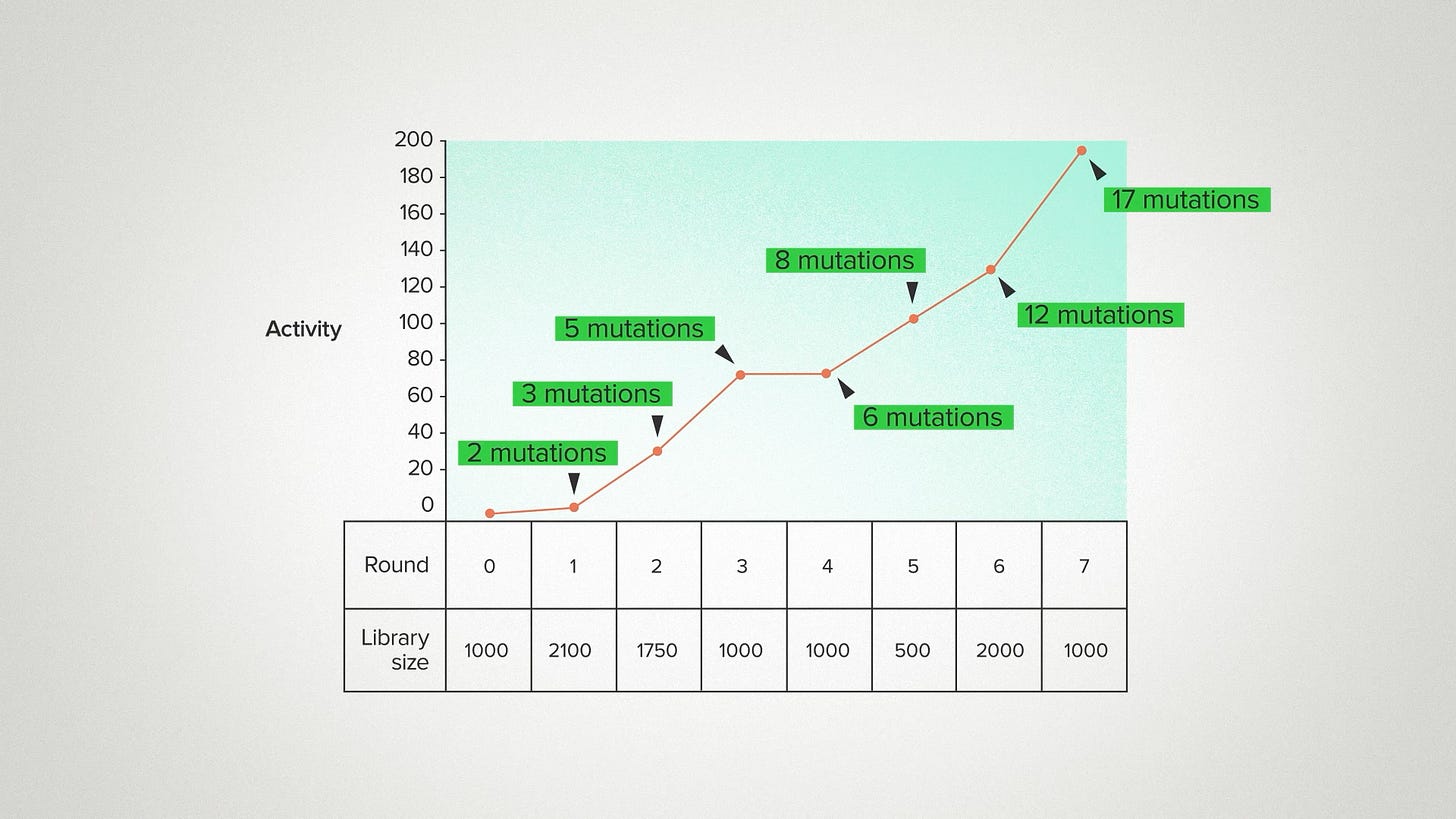
One way to understand what's going on here is to realize how much our ability to explore sequence space has changed. In the pre-AI era, we were usually constrained to exploring sequences very close to the original enzyme. We'd make two or three mutations, because there were no good tools to model sequences further away than that. You'd just be guessing at random. Or worse, you'd destabilize the enzyme and get garbage.
But with AI and other computational tools, we can take bigger leaps with more confidence. Here I'm showing the average number of sequence changes, relative to the wild type, for the best variants in each round: 3, 5, 12. The top performer in gen 7 was 17 mutations away from the natural sequence and the second best enzyme had 20 mutations - basically 4 parallel universes from where we started.
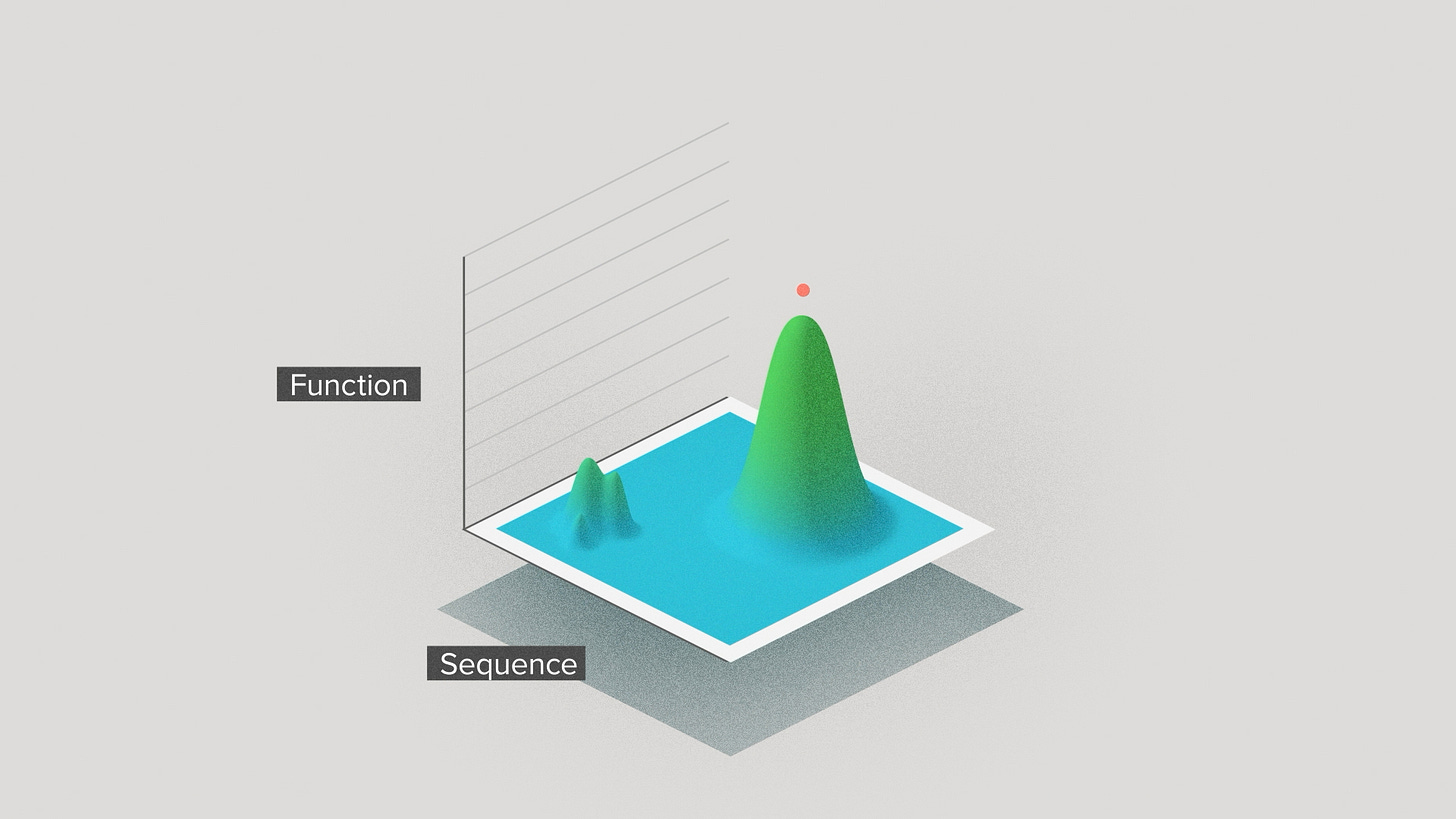
You can visualize this in terms of local versus global optimization on a performance landscape. Optimizing an enzyme is like climbing a mountain in sequence space. Traditionally, a project would start on the slope of a local mountain. Which mountain depended on the natural enzyme sequence you happened to have a tiny bit of characterization data for. You'd take small steps up the mountain and quickly reach the top.
But with AI, we can search a much wider range of natural sequences to discover more diverse starting enzymes. We can take bigger jumps that sometimes take us away from the local optimum and over to a different mountain entirely. We have a better chance of finding the highest mountain in the range - the true peak performance.
This perspective can help to reconcile the biological versus the AI philosophy to the world, the unstoppable force of AI versus the immovable object of hard problems. Biological systems still have limits. But, with AI, we can find limits that are global, or at least less local. So they may be farther away than we used to expect.





Could neuromorphic hardware take this even further, blending biological intuition with computational power? Can’t wait to see where this unstoppable force takes us next!
Fascinating! I’m curious how you think about the path dependence in this process. Surely the first 1000 sequences were a tiny sampling of the fitness landscape. Yet, those results probably helped design the next library (2100 variants) and so forth. Had you started with a different set of 1000 sequences in Round 0, how close would you have been to solution you ended up with?
Thanks for sharing these!