What is Synthetic Biology?
Synthetic biology is the evolution of genetic engineering: a toolset, a mindset, a community and a vibe. As an emerging technology, its boundaries are not fixed. But the field's guiding philosophy, bringing engineering approaches to biological problems, is a consistent source of inspiration and progress.
Transcript
What is synthetic biology? How is it different from genetic engineering? Today we're talking about why synbio is a thing and why it might be your thing. So come with me, fellow nerds, as I get lost in the weeds over semantics and we work out a definition for synthetic biology.
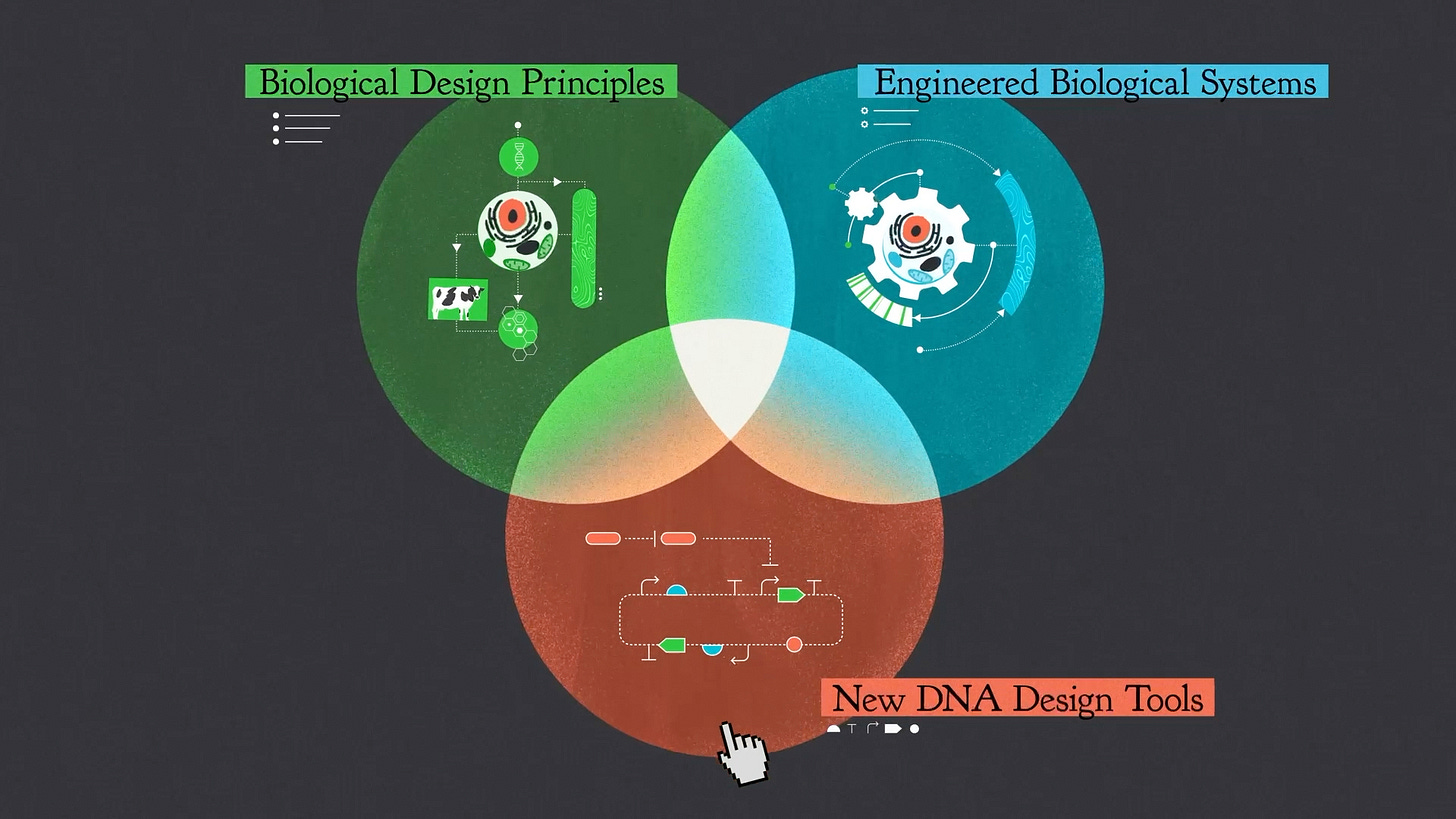
Let's get right to it. Synthetic biology is the engineering design process applied to DNA. A synthetic biologist is a scientist who seeks out design principles for biology - the rules that govern how DNA creates biological function. Or a synthetic biologist is a developer who uses those design principles to construct new biological systems that are interesting or useful. Or a synthetic biologist is an inventor who creates new technologies for reading, writing or editing DNA.
For everything synthetic biologists do, engineering DNA is the common goal. Now I hear some of you out there saying "Engineering DNA? Jake, I thought we already had a term for that, because that sure sounds like genetic engineering." And look - that's fair. I'm not here to be the word police.
But I do think looking at the different uses of these terms can give some historical and social perspective about what we're all trying to do here. So let's take a quick journey through time. The phrase "genetic engineering" became popular in the 1970s, just as the very first DNA technologies were coming online. It was an extremely productive era for biotech: picture the very first transgenic mouse, picture recombinant human insulin.
Those early pioneers had a very limited set of tools for working with DNA. Sequencing was still new tech, so they were usually working blind. DNA synthesis was barely possible, so they had to cut and paste existing genes. Measurement tools were pretty rough and involved things like eyeballing the sizes of spots on a blot. Laboratory automation didn't exist so they did everything by hand. Software? Forget about it.
In other words, early genetic engineering didn't have most of the features we associate with modern engineering: standardization, quantification, automation, simulation. It was more like an artisanal craft process, relying on human intuition and trial and error.
There wasn't any single breakthrough that transformed biology into an engineering discipline. By the '90s, you could sequence DNA routinely. By the 2000's, you could synthesize whatever you needed, at least for shorter pieces of DNA. Laboratory automation meant that you could standardize protocols and scale them up. Mass spectrometry, fluorescence imaging, and other measurement technologies made biological experiments more quantitative. Software tools let you model and simulate more biological complexity.
Sometime around the year 2000, a critical mass of people had noticed a pattern. Borrowing approaches from engineering and using them on biological problems was often a good idea. So they said, let's do that on purpose. Let's build biology around standard parts, quantitative measurements, computational tools and scalable automation. This is when "synthetic biology" became a thing.
The way that synthetic biologists passionately embrace engineering sometimes leads people to the understanding that synbio is like genetic engineering, only moreso. "Synthetic biology is genetic engineering on steroids" is one you might have heard. That's from an article in Science magazine in 2012. And I have to say I don't love it. I mean - do I look like I use steroids? But I get it. I respect the simplicity.
My version of a simple definition for synbio might go like this: "Synthetic biology puts the engineering in genetic engineering."
My extended definition for synbio is what happens when three things come together:
A tech stack of tools
A way of thinking
A community of people
The state of synthetic biology at any given moment is closely linked to the tools we have for reading, writing and editing DNA. Today that tech stack includes DNA sequencing, synthesis, gene editors like CRISPR, omics technologies, laboratory automation, and generative AI for DNA design.
The synthetic biology way of thinking is engineering-forward with a focus on enabling technology, measurement and the design process. If you've ever heard a biologist talk about the design-build-test-learn loop, the iterative process used for engineering R&D, that person was probably a synthetic biologist. They were engineering a yeast strain and they called it a good "chassis?" That's a synthetic biologist. They designed a piece of functional DNA and they called it a "genetic circuit?" Synthetic biologist. They were debating the similarities and differences between genetic code and computer code? Oh yeah.
In other words, we're huge nerds. And that brings me to our community of people. The most important reason why we need the term "synthetic biology" is that it helps bring people together. Fundamentally, synbio is what the synbio community does. The term will exist for as long as people want to bring the engineering design process to DNA, and want to find other people who think the same way.
If that sounds like a good idea to you then welcome to the club. I'm excited to hear about your favorite chassis for building genetic circuits.