The Bowtie Model and Complex R&D Workflows
An automated lab is like a metabolic network. No really hear me out, I think I'm onto something.
Transcript
There's a Venn diagram of people who love biology and people who love robots. I live right in the middle and I love it here. I'm never leaving. In this world, people will sometimes mix metaphors, looking to take inspiration from the world of robotics and apply it to biotech or vice versa. And there's one analogy in particular that I hear kind of a lot. It goes like this: R&D workflows should be more like metabolic networks.
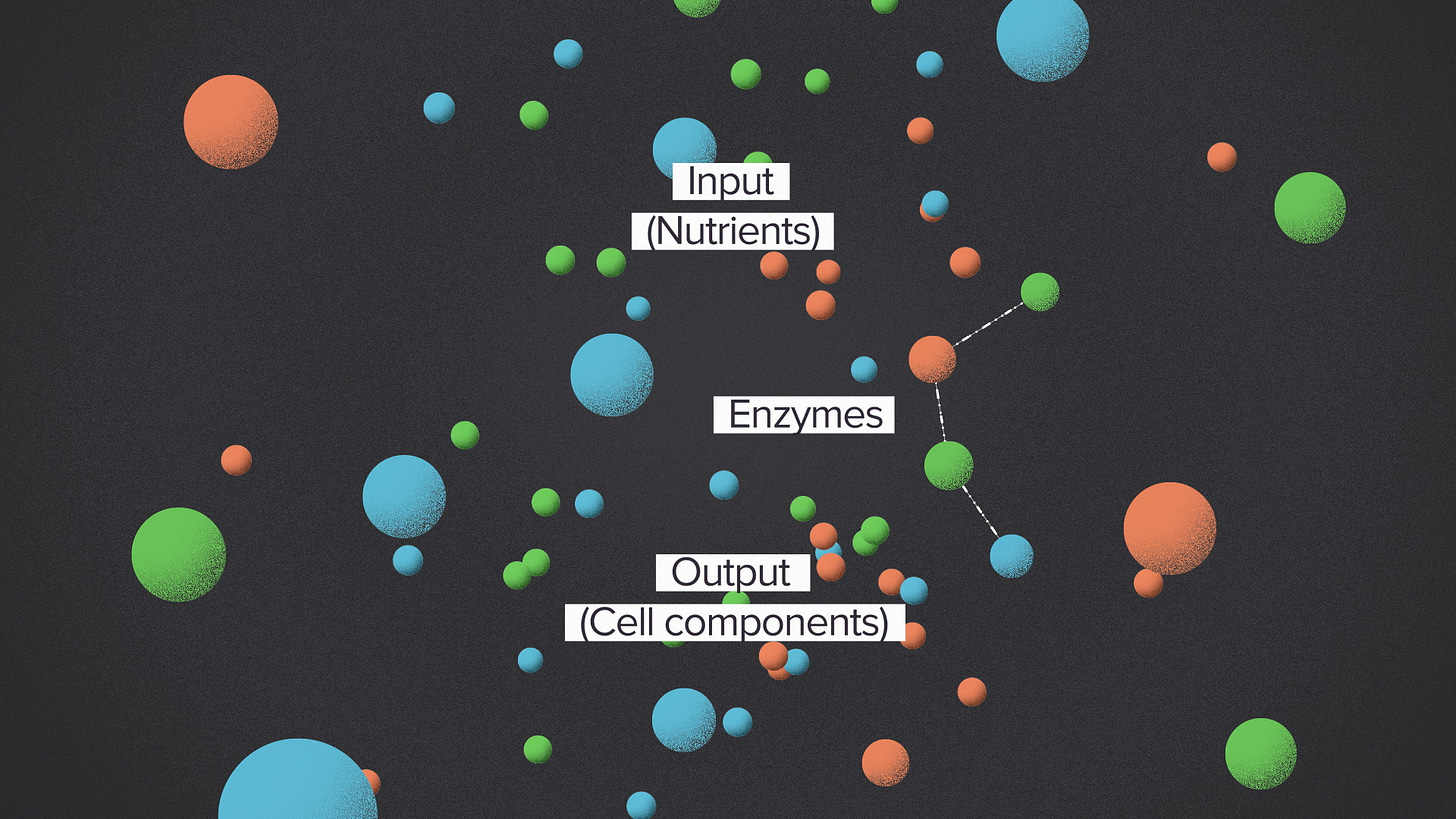
The inspiration comes from biology and how it organizes metabolism for the complex process of building a cell. At the highest level, metabolism is just a series of operations for converting inputs to outputs.
The inputs are the nutrients that a cell gets from the environment. A typical cell sees lots of molecules that are potential food. If it's made of carbon, something in biology will eat it.
The output that a cell wants to produce is more cells. Metabolism makes all the components that a cell needs to grow: hundreds of different kinds of molecules.
In between, the cell has enzymes. Each enzyme performs one chemical operation, transforming a molecule in a way that brings the inputs closer to the desired outputs.
So the structure of a metabolic network is a solution to a processing problem. Each species in nature has its own diet and its own cellular composition, so evolution is always tweaking and re-optimizing the solution. You could imagine a world where metabolism takes the direct approach, connecting inputs and outputs with the fewest number of steps. In this world, metabolism looks like a tangled network or a big cloud. Each organism's metabolic network evolves independently, a custom process with totally unique structure.
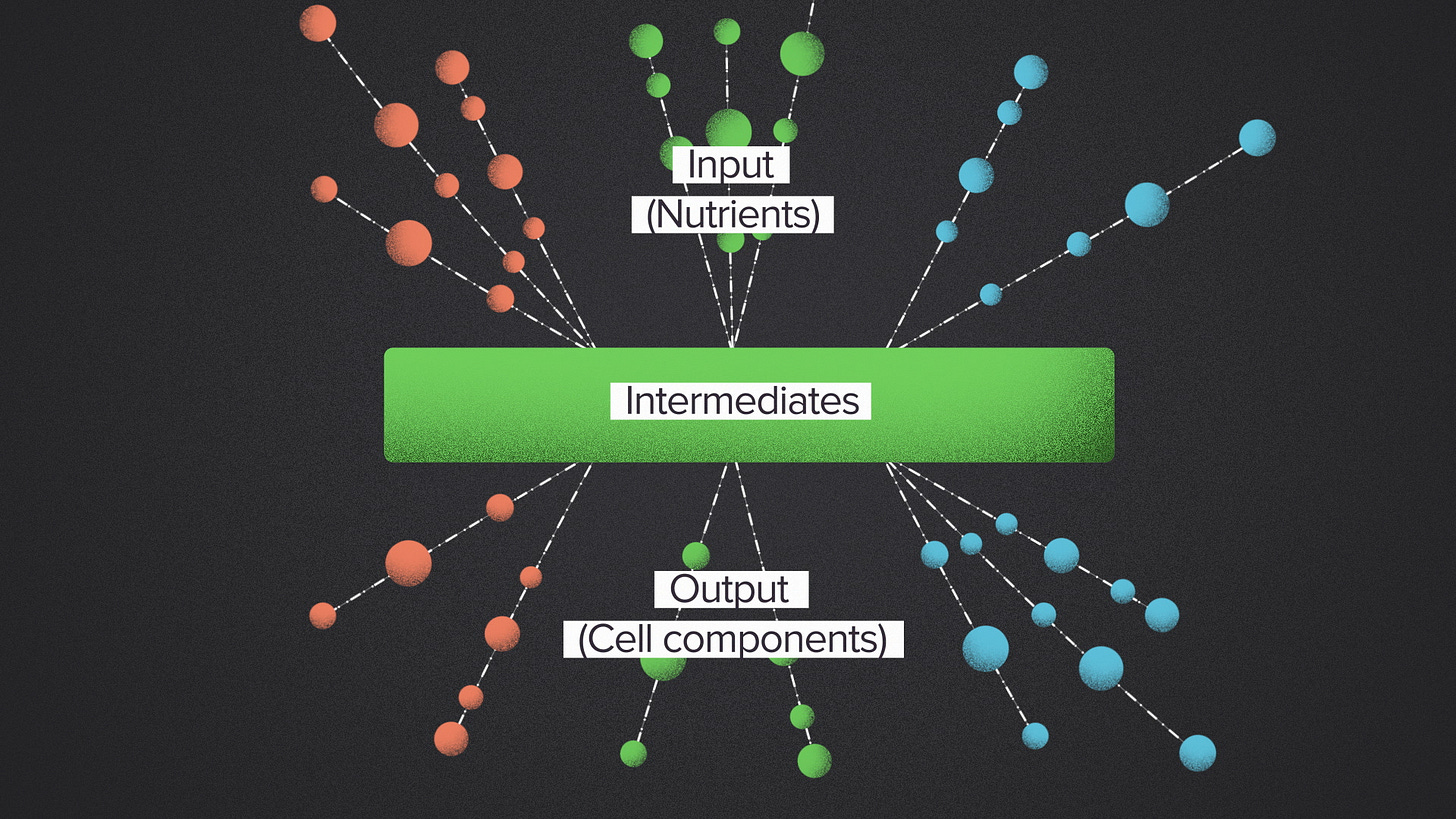
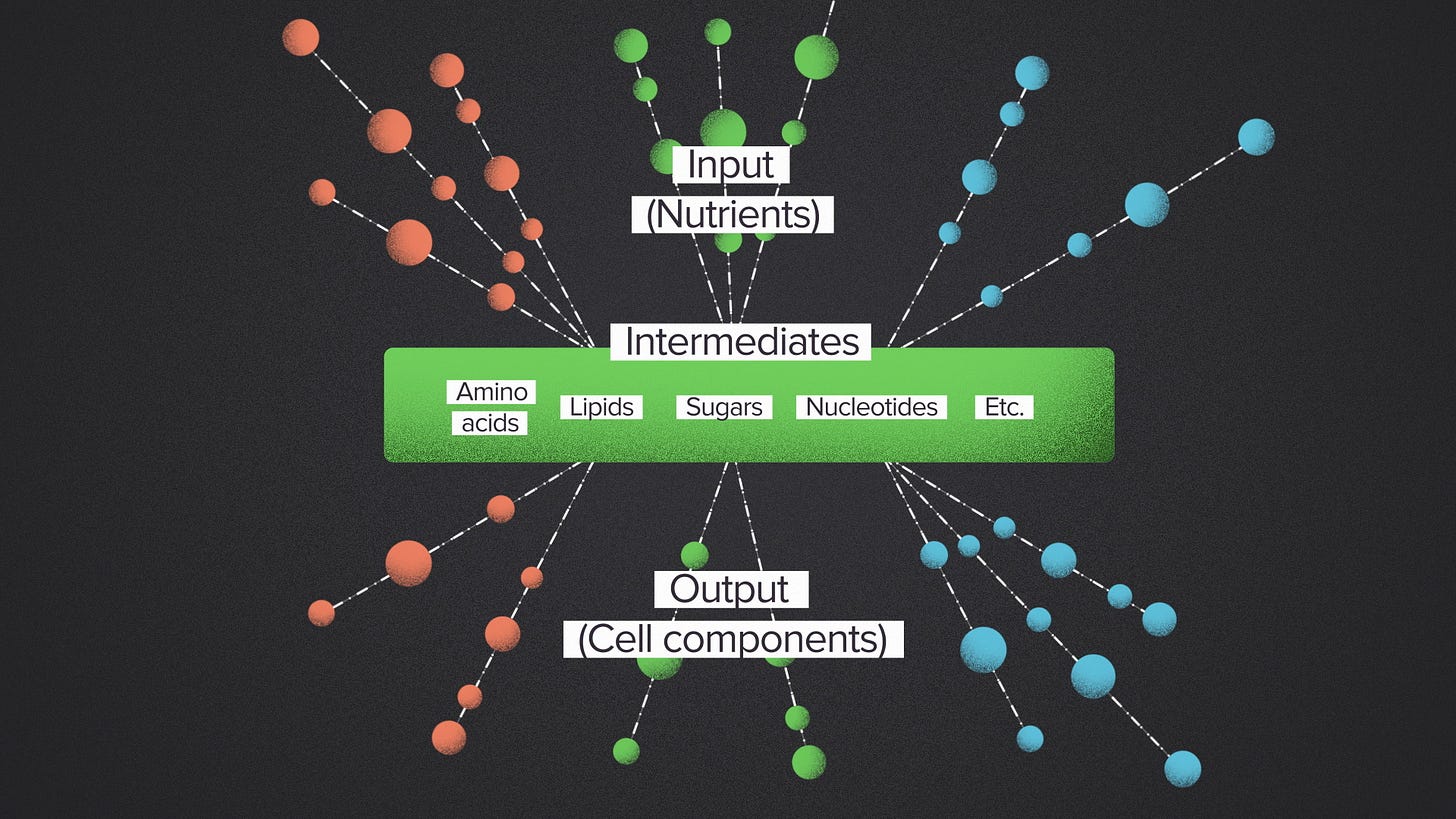
But that's not how nature does it. Instead, naturally occuring metabolic networks have what's called a bowtie structure. They're wide on top, narrow in the middle and wide on the bottom. The many input nutrients are broken down into a relatively small set of common intermediates. These intermediate building blocks are probably the most famous molecules of biology: sugars, lipids, amino acids, nucleotides. The metabolic outputs are built up using these building blocks as the source material.
Now, not every metabolic pathway follows this structure. This is biology, so of course there are exceptions. But it is remarkable how common the bowtie pattern is, and how stable it is across evolution. Every organism, as far as I know, solves the metabolic processing problem with a bowtie structure. It works for E. coli, the banana tree and the slow loris. It's the winningest workflow design in the history of evolution.
Why? I'd say it's because it enables flexibility and scale.
Bowties are flexible because they are modular. It doesn't matter how weird a given input molecule is. As long as the pathway can convert it into a standard intermediate, it can be used to build any kind of cell. That makes pathways more reusable. They can be mixed and matched to evolve new possibilities quickly.
Bowties can scale because cells can focus resources on processing just a few key intermediates. The ribosome, for example, assembles amino acids into proteins. For many cells, it is the key operation to enable fast growth. The cell can invest in building many ribosomes because they connect many inputs to many outputs. There will always be demand for more ribosomes.
Now that I'm primed to see this bowtie structure, I look for it everywhere. And I tend to find it in any complex process that can achieve both flexibility and scale.
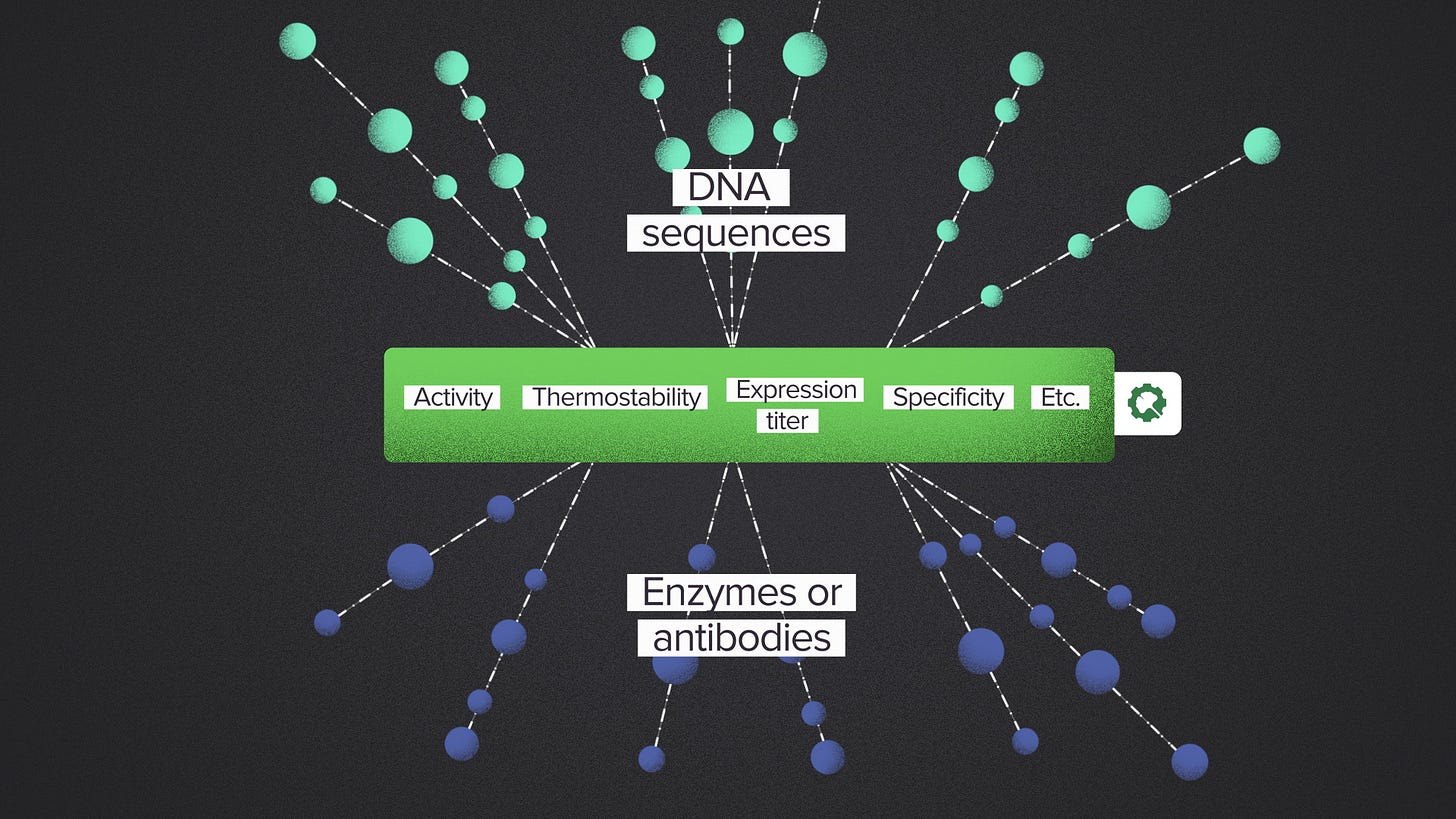
For example, here at Ginkgo, we offer lab data as a service. You can buy from us experimental data to characterize a biological product that you're developing, for example an enzyme or an antibody. These services have a wide range of possible inputs because you can specify the DNA sequence of your choice. And the outputs have a wide range of applications. An enzyme might be used to manufacture any number of chemicals or it could be a payload in a gene therapy.
But in between, there's a relatively small set of properties that are useful for almost all R&D projects: activity, specificity, thermostability, expression titer, etc. We can invest in the automation infrastructure to measure those properties at scale because there will always be more enzymes and antibodies to characterize. You get the flexibility to buy data for your application.
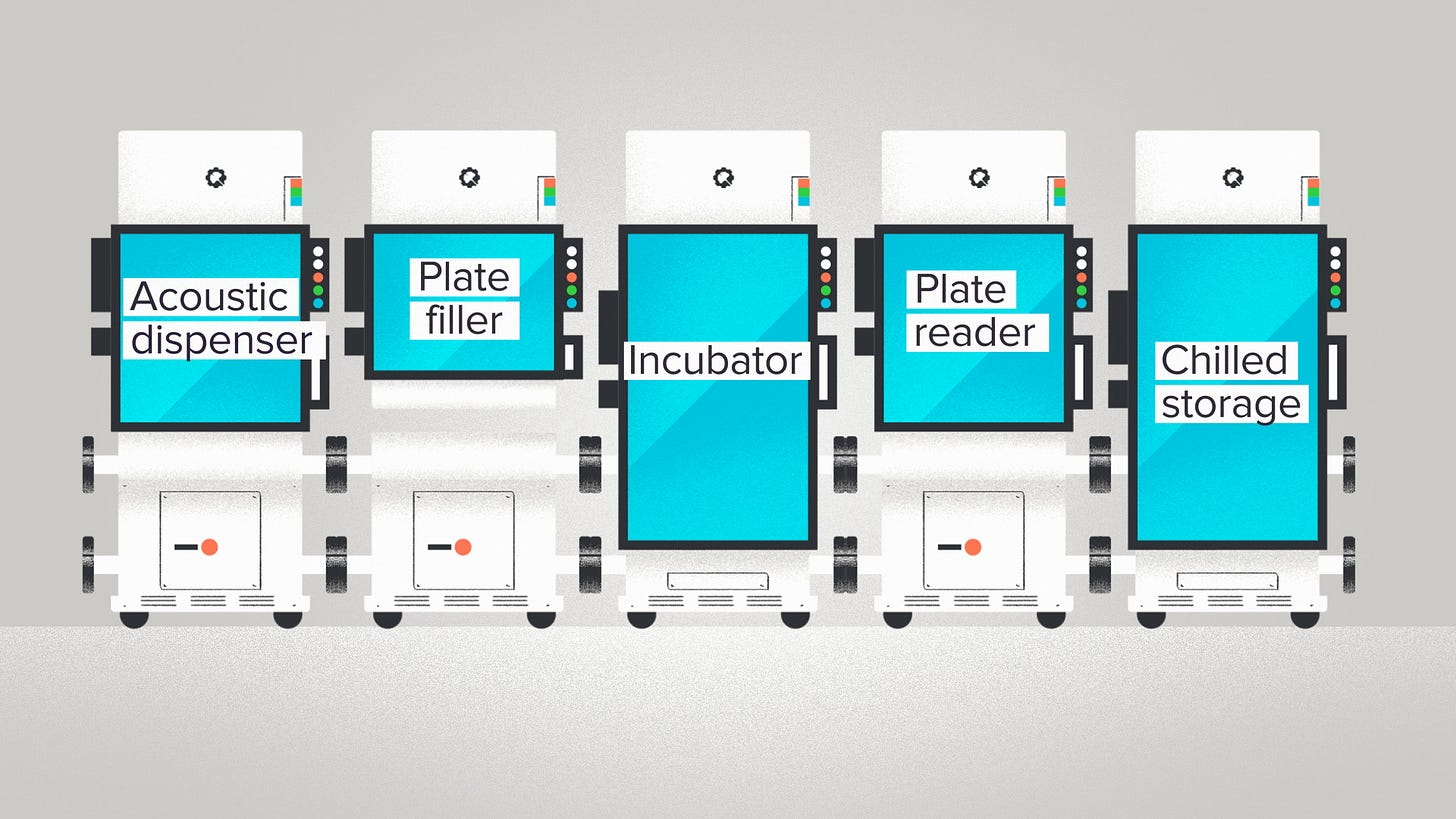
Or, another example. We organize automation workflows using RACs - Reconfigurable Automation Carts. In this case, the inputs are all the robotics that you'll find in a modern biotech lab: plate readers, liquid handlers, PCR machines, centrifuges, etc. The outputs are the entire universe of potential R&D workflows. Drug discovery. CRISPR screening. Sample prep for Next-Gen Sequencing. The RACs take all of those robots and package them into a few standard form factors, making them easy to connect together into a single assembly line.
The modular design makes RAC setups more flexible. You can change, reorder, or add new machines with a really light touch. It's a system built for rapid adaptation, that evolutionary advantage of a bowtie structure. If you want to try a RAC system with your lab automation workflows, they're for sale too by the way.
The success of the bowtie model of metabolism is still not completely understood. Why would so many organisms, across billions of years of evolution, converge on such a fundamentally similar solution to such a complex problem? Those of us who love biology want to understand the structure of metabolism. And those of us who love automated biotech R&D want to copy the structure of metabolism.