The Biological Codebase at Ginkgo Bioworks
Today we're talking about the biological codebase as a resource for engineering biology with flexibility and scale.
Transcript
Open up a biology textbook to chapter 1. There's a good chance it talks about the unity and diversity of life. Because we share a common ancestor, living things share common properties: DNA, cell structure, metabolism, etc. These common properties are like a set of building blocks from which evolution generates variety: yeast, cabbages, slow lorises, what have you.
Unity and diversity are how biology is organized, so they should translate to the way we engineer biology. Conceptually, biology can do a lot of different things: Gene therapy, plant traits, industrial biochemistry. So some aspects of our engineering strategies will be different for every project. But biology is also unified, so there should be some things that we can standardize and replicate across many projects. We need flexibility and scale.
Most biotech R&D projects today don't benefit from scale. They are organized as if each biological system was completely unique. A team comes together around just one product, they design everything from scratch, they do the work by hand or in small batches of automation, and when the project is done, nothing is saved for reuse.
At the other end of the spectrum, most big automation facilities out there aren't flexible. They're purpose-built for just one purpose. A complete biotech R&D project needs many different kinds of operations, so scaling just one of them can be pretty marginal. Because biotech is risky, it's hard to justify big investments for scaling just one project.
So how does the Ginkgo foundry do it? How do you create one facility that can execute many different projects for many different partners, with each of them being able to benefit from scale? The answer is the biological codebase.
At Ginkgo, we use the term codebase to refer to anything about engineering biology that is reusable or repeatable across many projects. It's a term we borrow from software development, where they know the value of building with reusable chunks of code. Ask an iOS or an Android developer to write every line from scratch? They won't even understand what you're asking.
Biology lives in the world of atoms, not bits. So the biological codebase is more complex than a library of computer code. It includes assets, things that we have, and capabilities, things that we can do. It includes physical things like cell lines and automation hardware, and digital things like data for training AI models. Personally, I'd break down the biological codebase into 6 different categories.
Chassis organisms are the strains and cell lines that we work with the most. Yeast strains like Pichia or mammalian cells like Hek. We don't limit ourselves to only one cell type, but the more you work with a particular organism, the better you get at all the techniques required to engineer it.
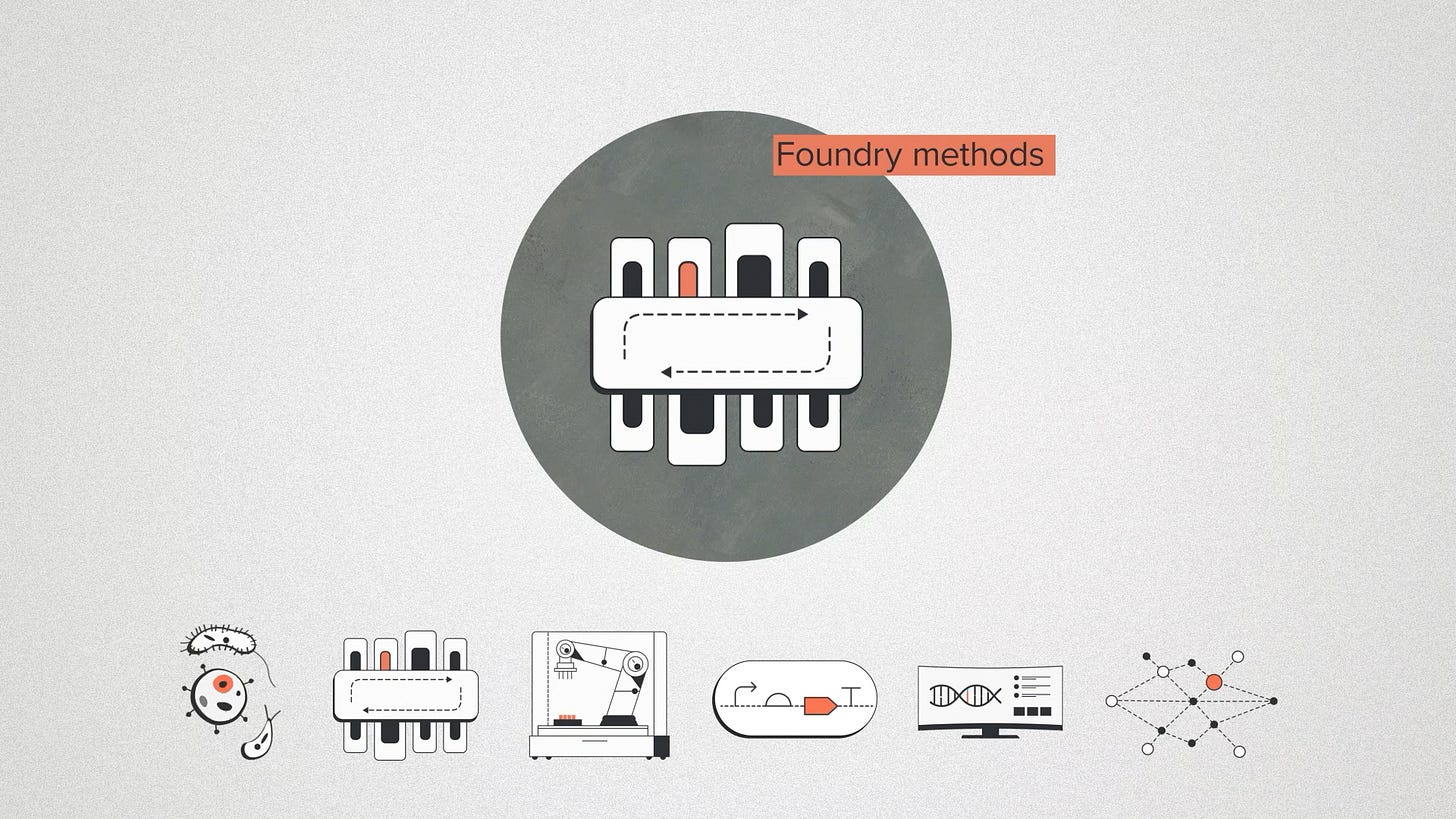
Foundry methods include protocols, assays, and everything you need to know to work with biology effectively at scale. How do we grow these cells? How do we get DNA into them or edit their genomes? When methods are reusable, they can be iteratively improved and refined. The more methods we onboard, the better visibility we have on the right approach for a given problem.
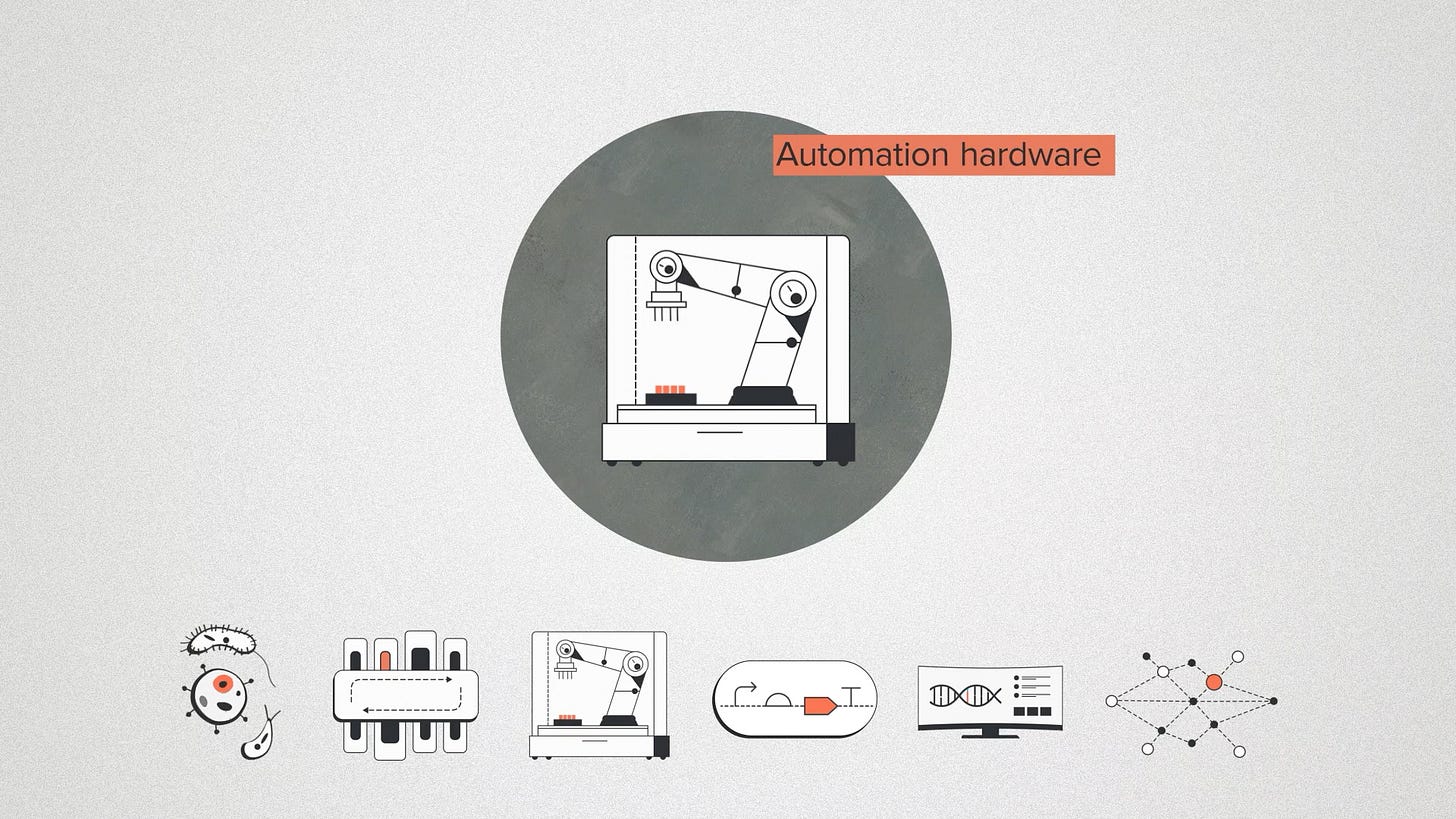
Automation hardware is all of the physical infrastructure that we use to perform those methods. This includes hardware that's familiar to most biologists, like liquid-handling robots and plate readers. It also includes custom robotics like the RACs: Reconfigurable Automation Carts that can be mixed and matched into a specific workflow as the project requires.
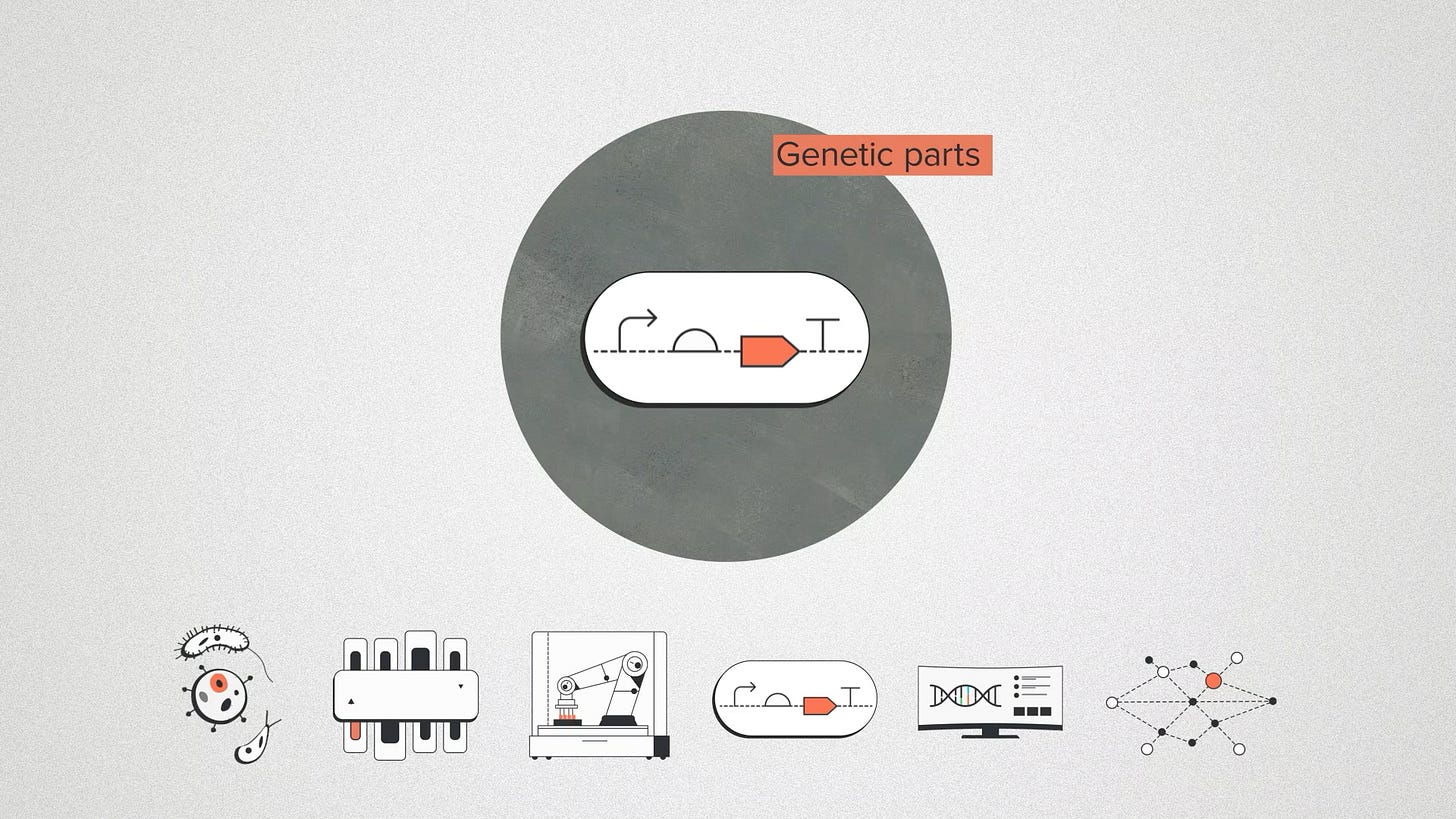
Genetic parts are pieces of DNA that perform a commonly needed function. Usually these are not the core function of your DNA design, but the nuts and bolts that make it work. Think about strong promoters for making a lot of protein, think about vectors for carrying DNA to the right place. Control elements. Signaling domains. Gene editors.
DNA libraries are a resource for discovering new pieces of functional DNA. Our big one is called the UMDB for Unified Metagenomic Database. As of this recording, it has more than 2 billion unique genes. We can search this for previously unknown and uncharacterized sequences, like a protein or an enzyme, that perform an essential function in your design.
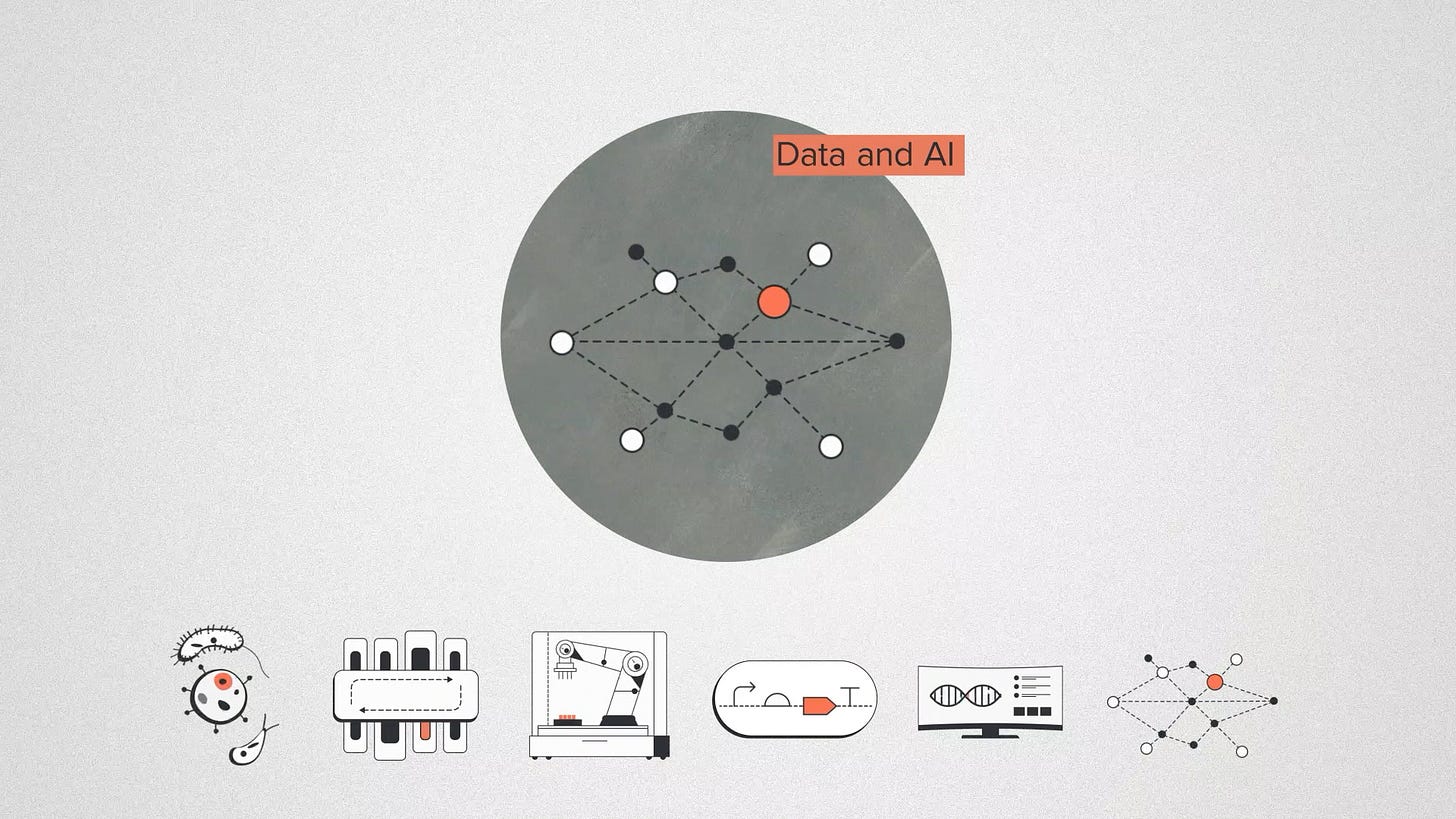
Finally Data and AI I'll group together, because they work together to be the fastest growing part of the codebase. In the foundry, we produce tons of biological data. AI can be trained on these large scale datasets and it has the flexibility to generate solutions for a specific problem.
It can be hard to summarize the biological Codebase because of its multiplex nature. But that's how it has to be. Codebase only works because it is a collection of assets and capabilities that are diverse enough to span real biotech R&D use cases.
A typical project taps into the codebase multiple times. Let's say you need an enzyme for manufacturing a small-molecule pharmaceutical. We can search our DNA libraries for enzyme candidates, use generative AI to refine and optimize their sequences, add some off-the-shelf genetic parts to express those enzymes at a high level, then transform the DNA into a chassis organism that is suitable for manufacturing them at scale.
Your enzyme is designed uniquely for your application, but all this other stuff that is supporting the R&D is reusable. You're benefiting from the fact that we've used all these tools before and that we didn't have to build out these unique capabilities just for one project.
Codebase is the right way to organize biotech R&D. Software developers don't start new projects from scratch. Evolution doesn't generate novelty from scratch. They both mix and match existing components, then adapt them to specific conditions. When bio and tech both agree on the right way to develop complex systems, it's probably a good idea.