Evolved vs. Engineered Proteins in the AA-0 Model
Sequence-based LLMs learn patterns from proteins found in nature. So what happens when you want to engineer a function that's not found in nature? Today we're looking at some results from AA-0, the first model on Ginkgo's new AI developer platform.
Transcript
To build anything with biology, first you've got to learn from the best: nature. In the arts, people talk about the "anxiety of influence". A painter compares themselves to the great artists of the past and thinks "Wow - will I ever make something that good?"
It's like that for synthetic biologists. Except our great artist is evolution, the thing that made all life on earth. Like - have you seen trees? They're amazing. That's a tough act to follow. We can all agree. That's hard to compete with.
Lucky for us, nature doesn't mind when we copy ideas. So engineering biology has mostly been about remixing parts from nature. We take a piece of DNA from one organism and move it somewhere else. Maybe we change a few base pairs to tweak the function.
And this works well for us because nature provides so many functions to work with. Transcription factors, enzymes, CRISPR arrays - nature's toolbox has all the parts we need. Or so I thought. Now I'm not so sure. Lately I'm wondering if some key functions are not going to be things we can just remix from nature. What changed my thinking is AI.
The Ginkgo AI team recently released a new model called AA-0, for Amino Acid Zero. You can play with it on our AI portal - I'll put the link below:
It's built similarly to models like ESM-2 that have done very cool stuff for protein engineering. The difference is that this one is trained with our in-house database - about 2 billion sequences that are mostly not found in the public repositories.
I'll do a little background for people who are new to protein LLMs. Their training data is a large set of naturally occurring protein sequences. They learn the patterns that influence how likely a particular amino acid is to appear at a particular position. So you can use them to score a protein sequence for how much it resembles, in a statistical sense, a natural sequence. Or you can generate new sequences that look naturally evolved.
Now, as protein engineers, we're not trying to make natural proteins. That's the whole point. But being able to score a sequence for how "natural" it looks is still useful. Because naturally occurring proteins have passed through the filter of evolution, they probably have some function. They do basic things, like fold into a stable shape, that an engineered protein also needs to do. So there's reason to expect that proteins that score high on being evolution-like will tend to make good engineered proteins.
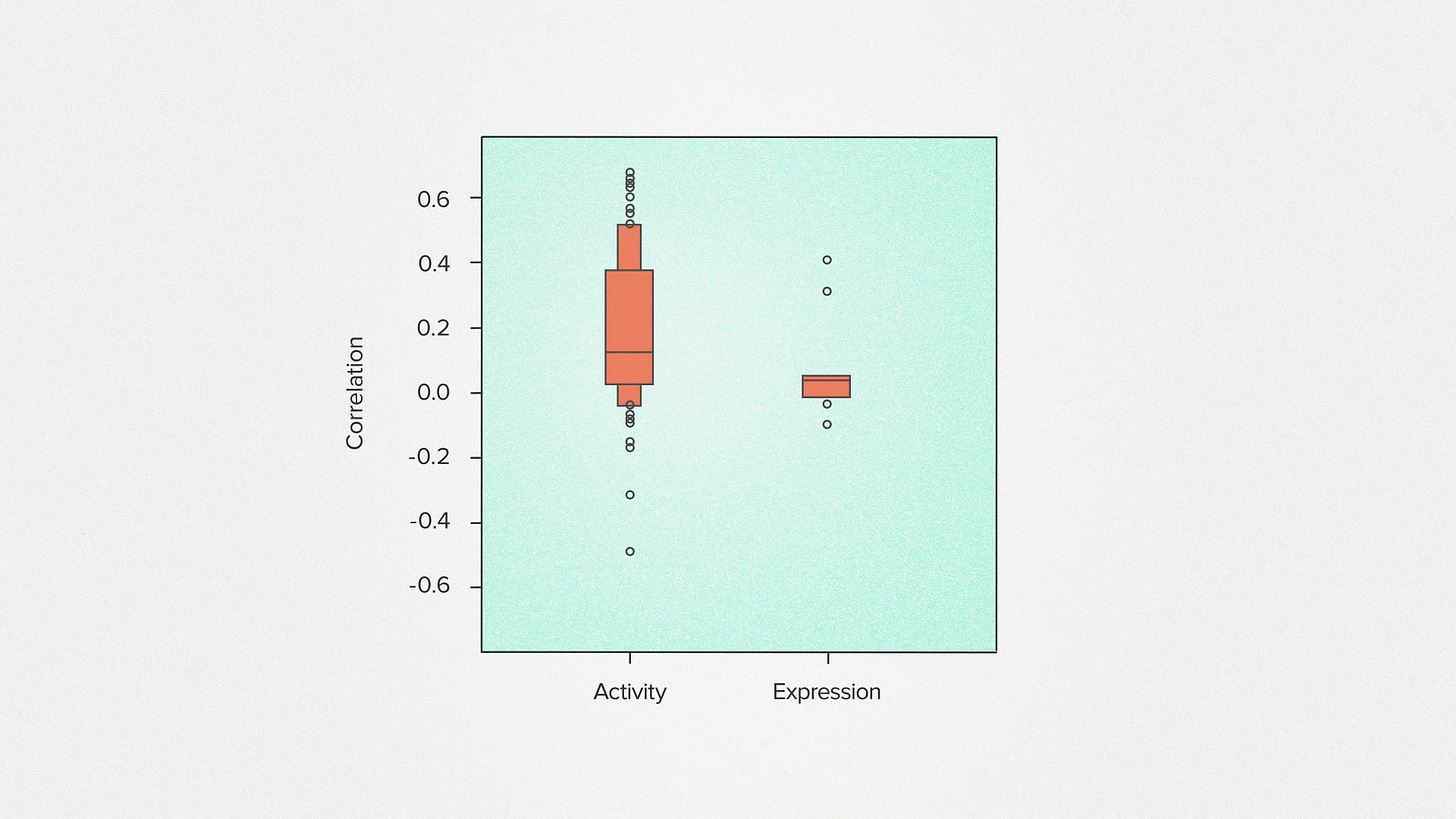
We wanted to try this out for some protein engineering tasks we do here at Ginkgo for our customers. So we put together a panel of 73 different tests. Each test includes a collection of protein sequences and, for each sequence, a measurement of a functional property. For example we might measure activity - how fast does the enzyme work? Expression - how much of a protein is being made. Or specificity - how selective is it for a particular molecule.
We compare the measurements to the model-generated scores and we get a correlation. We can think of this as a relationship between how natural a protein looks and how functional it is. I'm going to collect the results from different tests into a box blot to give an overall picture for how the model performs.
So how did AA-0 do? For measurements of activity, we get mostly positive correlations. Fast enzymes tend to resemble evolved enzymes. This is what I would have expected and similar to results that have been found for other models.
For measurements of expression, we get basically no correlation. Sequences that look more evolution-like aren't being made by the cell at higher levels. This seems reasonable to me because I know that a lot of the control of protein expression happens outside of the protein sequence itself, in other parts of the genome that the model doesn't see.
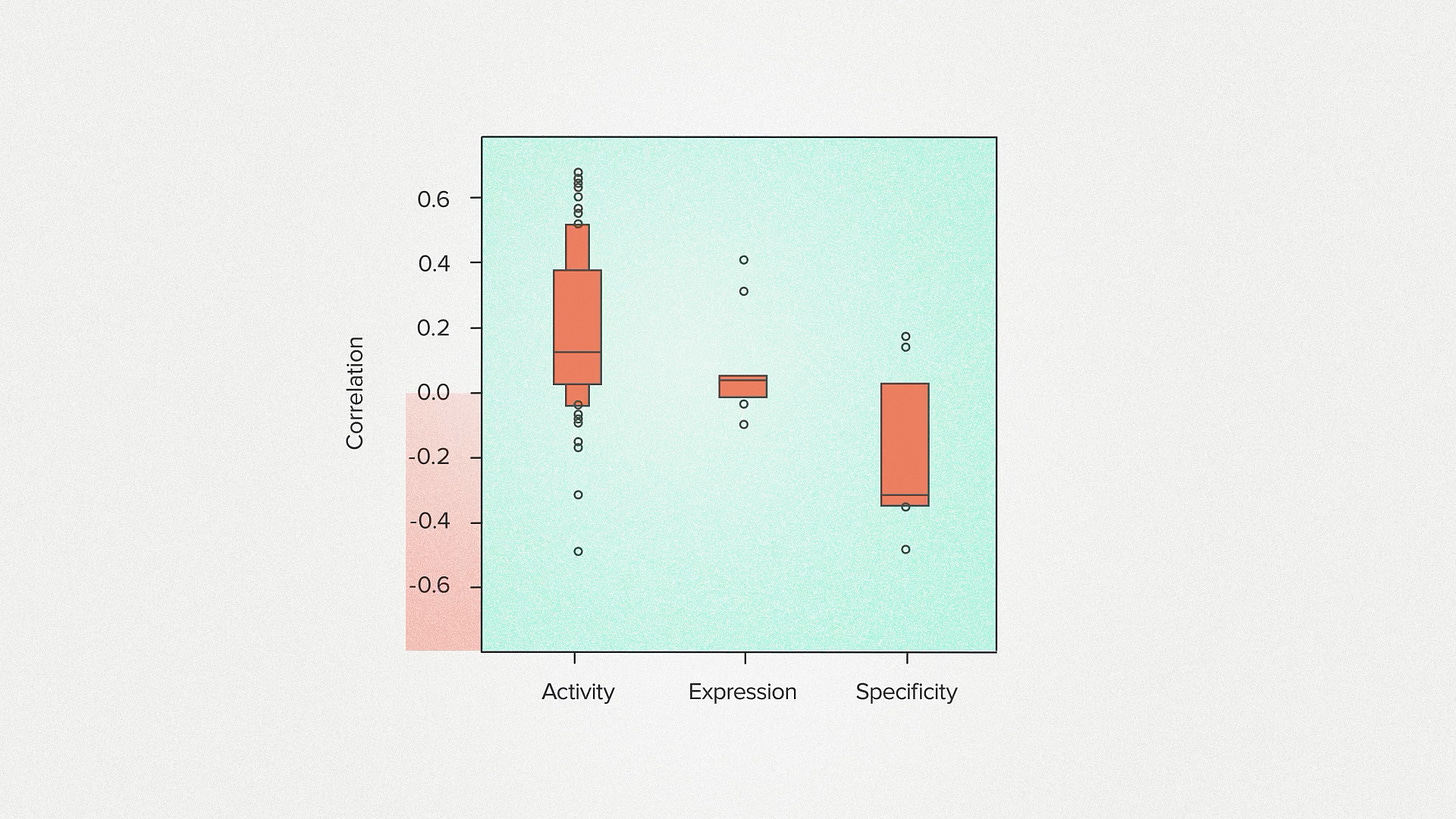
But now look at specificity. It's the complete opposite of what a protein engineer would want. For many enzyme projects, we tend to get a negative correlation. Enzymes with better actual performance have worse model scores. What's going on here? Does evolution hate specificity?
Now, this is preliminary. It hasn't been peer reviewed. We need more data. But I want to talk about it anyway because it made me reconsider the relationship between engineering and evolution. It suggests that, at least sometimes, they can disagree. The amino acids that nature would tend to use are the opposite of what worked for us. Why?
Maybe it's because of the enzymes we work on. Engineering isn't easy. We do it because we couldn't find the solution already in nature. So there's a kind of selection bias where engineering is taking on the jobs that evolution rejected for whatever reason.
Another possibility is that evolution doesn't value specificity as much as we do. Nature usually isn't trying to manufacture one clean molecule. Evolution might favor versatility over precision. If we want a function that evolution doesn't, our sequences are going to score low on the scale of evolution-likeness.
Or maybe it's something else that I haven't thought of. But I can't help but wonder if this means there might be functions out there that aren't in nature's toolbox. We can't copy them directly from natural sequences and we can't AI-generate them using only natural training data. We'll need to train AI models on engineered protein sequences. If the 2 billion sequences that went into AA-0 are any indication, we're going to need a lot of this data if we want powerful AI capabilities specific to engineered, rather than evolved, sequences.
Don't get me wrong - nature is still the old master. Almost everything we do as synthetic biologists is copied from nature or inspired by nature. But this is a moment where we might take a tiny step toward doing our own thing and that seems like a big deal to me.
For more on AA-0, check out this recent technical report on the Ginkgo website:
AminoAcid-0 (AA-0) A Protein Sequence LLM Trained on 2 Billion Proprietary Sequences