Engineering biology is sometimes a numbers game: the more biological designs you can build and test efficiently, the better your chance to find that superstar strain. This is especially true for strategies that seek to improve strains through unbiased mutation and selection - without the application of targeted genetic engineering.
EncapS is Ginkgo's technology for Encapsulation and Screening in ultra-high throughput. By packaging microbes into nanoliter-scale bioreactors, we can quickly screen for high performance microbes in batches of 100,000 at a time.
Transcript
Let's talk about a cool piece of technology we've got here in the foundry called EncapS, for encapsulation and screening. EncapS allows us to select for high-performing microbes 100,000 at a time.
You may remember artificial selection as the process that gave us dogs and corn. It starts with the genetic variation that happens naturally in every population. Humans look out for traits that they prefer and then select those traits to grow and propagate. It's evolution, but with humans guiding it on purpose.
We can do selection on microbes too when we're looking to cultivate desirable traits - the microscopic version of dogs. And selection can be a powerful strategy for microbes because of their large numbers and diversity.
To make this work, we need a way to recognize when a microbe has a desirable trait. In other words, we need to perform a measurement. We need a way to separate microbes on the basis of that measurement. And we need to do this at scale in order to effectively sample the diversity that exists in large microbial populations.
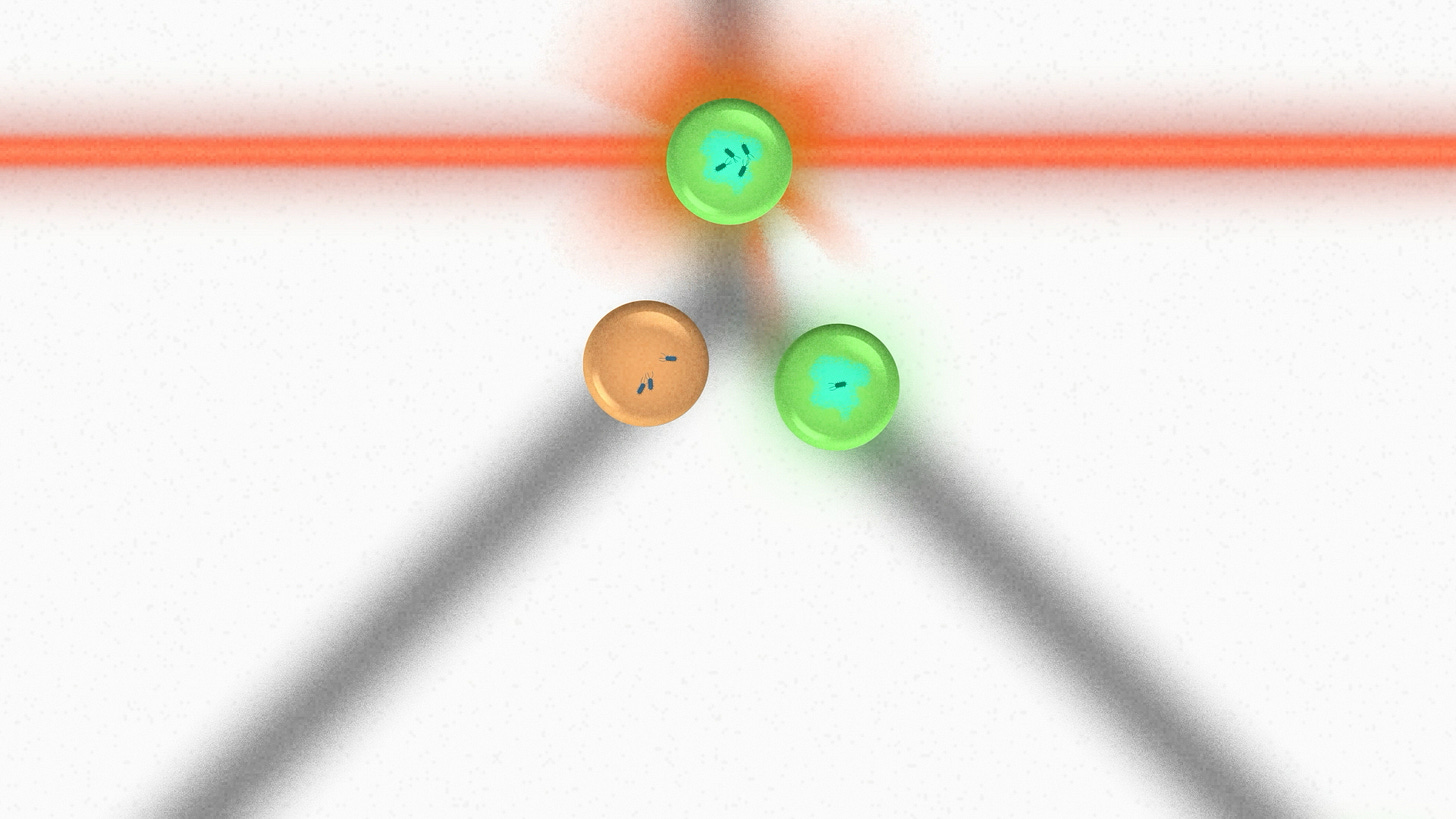
At Ginkgo, we automate this process with EncapS. Here's how it works. We start with a microbial culture. We use natural mutagenesis or some other strategy to create diversity in this population. Then we encapsulate the microbes into tiny little hydrogel beads. We call them nanoliter-scale bioreactors - you can think of them as very small test tubes. Each hydrogel bead includes a little bit of media and a little bit of room to grow.
As the microbes grow in these nanoliter-scale bioreactors, they can secrete proteins or make fermentation products, all of which are captured inside the bead. After an incubation period, we use lasers to perform a fluorescence measurement on the beads and sort them according to the results. From a large initial population, we collect a small number of top performers.
You might have heard of other methods that use lasers to sort out individual bacteria like FACS (Fluorescence-Activated Cell Sorting). The key difference here is that FACS doesn't package cells with their surrounding media, but EncapS does. That means EncapS can allow us to select for traits that go beyond the cell itself and affect the surrounding environment.
What kinds of traits can we measure with EncapS? Growth. Fermentation products. Protein production. Protein binding. Cell-cell interactions. In principle, we can select for something with EncapS if we have a way to measure it using fluorescence. Fluorescence-based assays are pretty common, so for some applications we find that one already exists. In other cases, we can develop an assay that works in the nanoliter-scale format. We have a pretty versatile toolbox for doing EncapS measurements on complex phenotypes and we're coming up with new stuff all the time.
I saw a cool example of this recently that involves co-encapsulation. We can mix more than one different cell type and perform measurements based on their interaction. In this case we were selecting for microbes that produce an antibiotic, so we mixed them with microbes that are targeted by the antibiotic. We labeled the target microbes with GFP to produce a fluorescence signal, then selected for beads where the signal dropped.
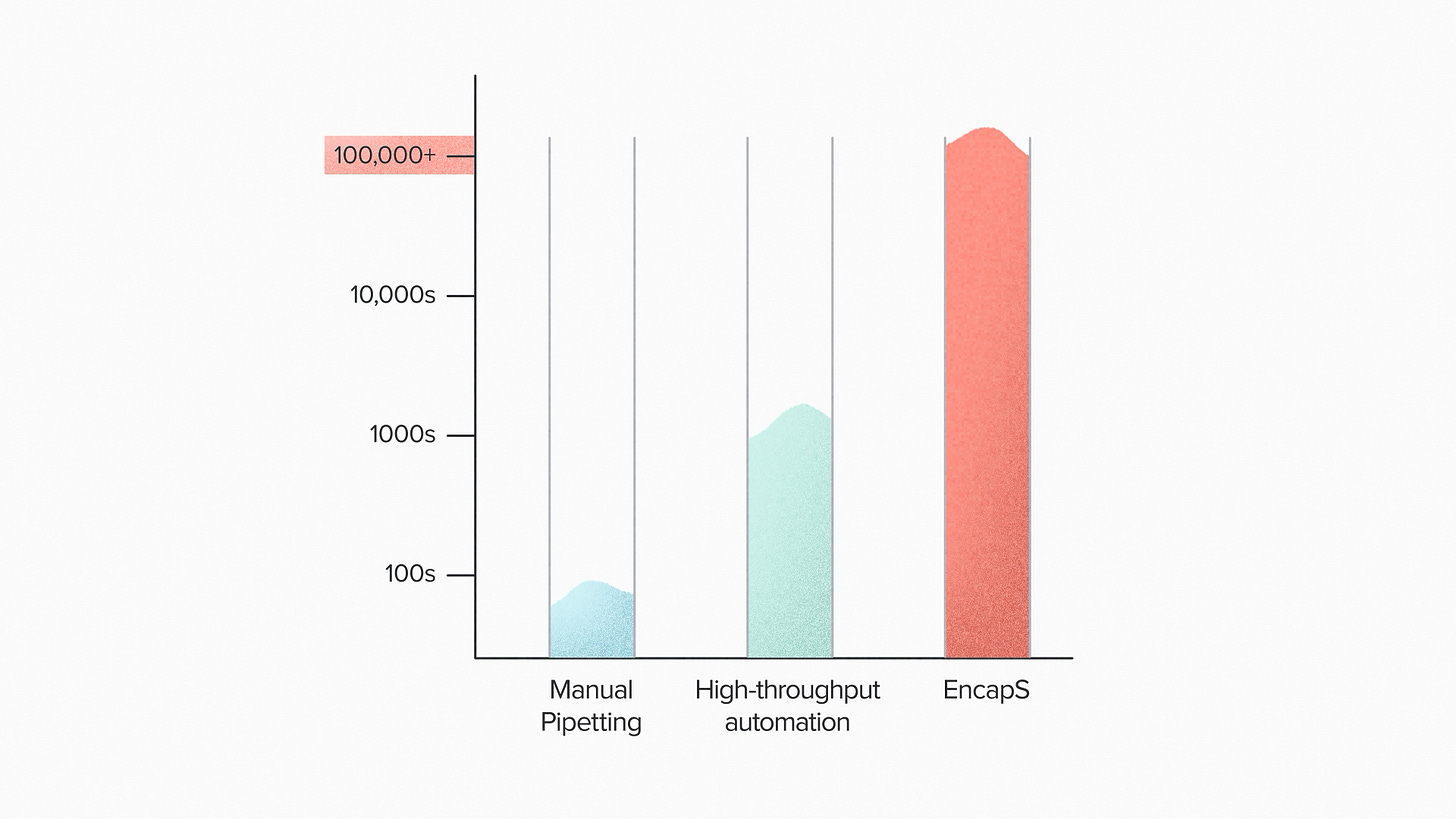
The power of the EncapS system is in the sheer numbers of beads it can screen. A human scientist could test maybe hundreds of microbes. With conventional lab automation, that number goes to thousands or tens of thousands. With a system like this, we can screen 100,000 or more microbes in a single run. Microbial selection is very much about sheer numbers, because we're looking for those rare high-performing superstars.
What kind of R&D projects are good candidates for EncapS? First of all, EncapS is a powerful option for those cases when you can't or don't want to do genetic engineering. For example, you might want to optimize a microbe that will be used for agriculture or for the human microbiome.
Second, EncapS is a great complement to genetic engineering. Genetic engineering is about making focused and intentional changes to a microbial genome. Selection is about trying everything in an unbiased way and seeing what works. We don't know everything about how to optimize performance in a single cell. So EncapS can boost performance on top of a rational synthetic biology campaign.
Finally, I would consider EncapS for unusual microbes that are more difficult to engineer directly. The encapsulation approach is versatile and works with different cell types. We can do bacteria, yeast, fungi or even mammalian cells. So there's a good chance it will work for that wild-type strain or unusual cell type that no one has ever worked with before.
As a biologist, I find EncapS to be amazing. Screening 100,000 or more nanoliter-scale bioreactors at once is mind blowing to me. Natural selection is the most powerful creative force on earth. EncapS, puts a little bit of that power into our process for developing more productive and beneficial microbes.