A Cleaner Interface for Biotech R&D
What is the structure of biotech R&D? Can we break it down into pieces? Should we even try? Today we're looking at different stages of biotech R&D and where they connect to the Ginkgo foundry.
Transcript
Biologists tend to be holistic thinkers. An organism is an integrated thing. The foot bone's connected to the leg bone. The MAP kinase is connected to the PI3 kinase. We like to keep track of all possible connections because you never know what might be important.
This philosophy carries over into how we design R&D projects. We try to maximize the integration of planning and execution. Any given detail in step 7 might influence a choice we make at step 3, so we need to keep the whole thing in our head and keep the whole team in constant contact.
Contrast this to fields of engineering where work is more subdivided and structured. Hardware vs. software. Operating system vs. application. Front-end vs. back end. A technology product is also an integrated whole - everything on a smartphone has to work together - but the work is split up into functional pieces that interact through specific channels.
The cost of this is that nobody gets complete control over everything. The web designer doesn't get access to the server's machine code. But the payoff is that each layer in the tech stack can be more focused and efficient.
So - can we do this for biology? In the case of engineering DNA, we seem to have an analogous challenge. A living thing is a complex system written in digital code that can, in principle, do almost anything. We could centralize the decision making for every base pair. Or we could distribute the work across more specialized sub-routines.
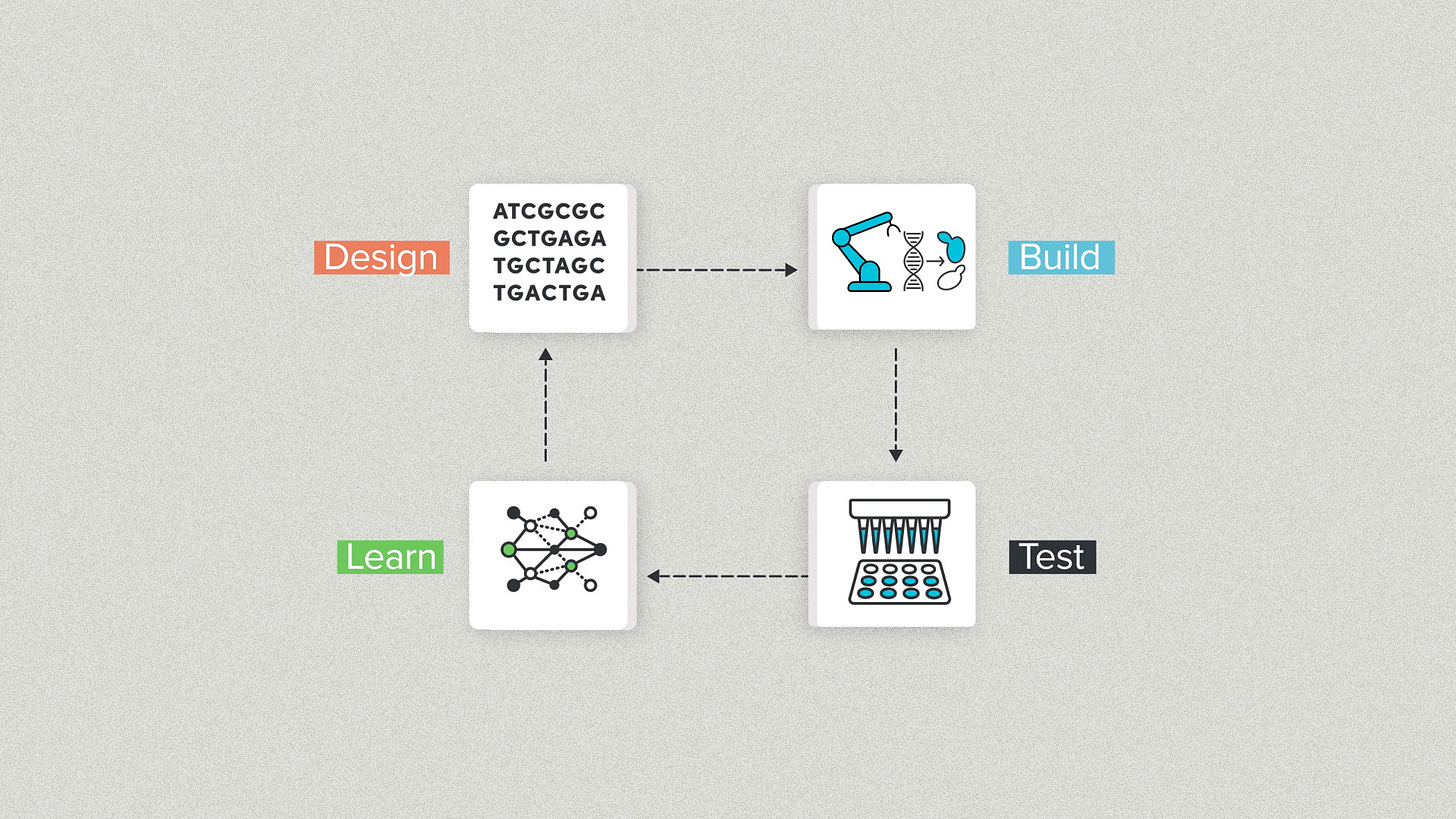
One popular schema is the DBTL cycle: Design, Build, Test, Learn. Synthetic biology stole this from classical engineering but it works for DNA too.
Design is writing DNA sequences.
Build is physically making that DNA in the real world and getting it into a living cell.
Test means taking experimental measurements to evaluate the design's performance.
Learn is analyzing the data and making improvements.
At Ginkgo, we often use DBTL internally because it helps to divide complex projects among specialized teams. The payoff is simpler and more streamlined communication. Design can pass digital DNA files to Build. Build can pass physical DNA to Test. Test can pass experimental data to Learn.
As we get better at this, the inputs become clearly defined and calling a service requires less communication outside those channels. Eventually the service calls get clean enough that it starts to make sense to offer them externally too.
For example, Design. With the Ginkgo AI portal, you can use our AI models to generate your protein sequences. The AA-0 model was trained on about 2 billion proprietary DNA sequences that we've collected for other applications. The work of building that dataset happens in the Ginkgo backend. All you need to access it is a simple API call.
Or consider Build and Test. You want to design the DNA sequences, then get functional data as output. That works. If you're making an antibody, you can get antibody developability data from the Datapoints team. The lab automation infrastructure lives in the Ginkgo backend. You access it with a fee-for-service deal.
Or how about Design and Build? Maybe you want to do the testing yourself. For example, you're doing pharmaceutical manufacturing and you need enzymes to perform in your existing process. We offer sample plates of pre-designed enzymes with popular chemistry. You can evaluate them for free for your process and, if you like the results, license the enzyme from us. Or we'll put together a custom enzyme plate for a small fee.
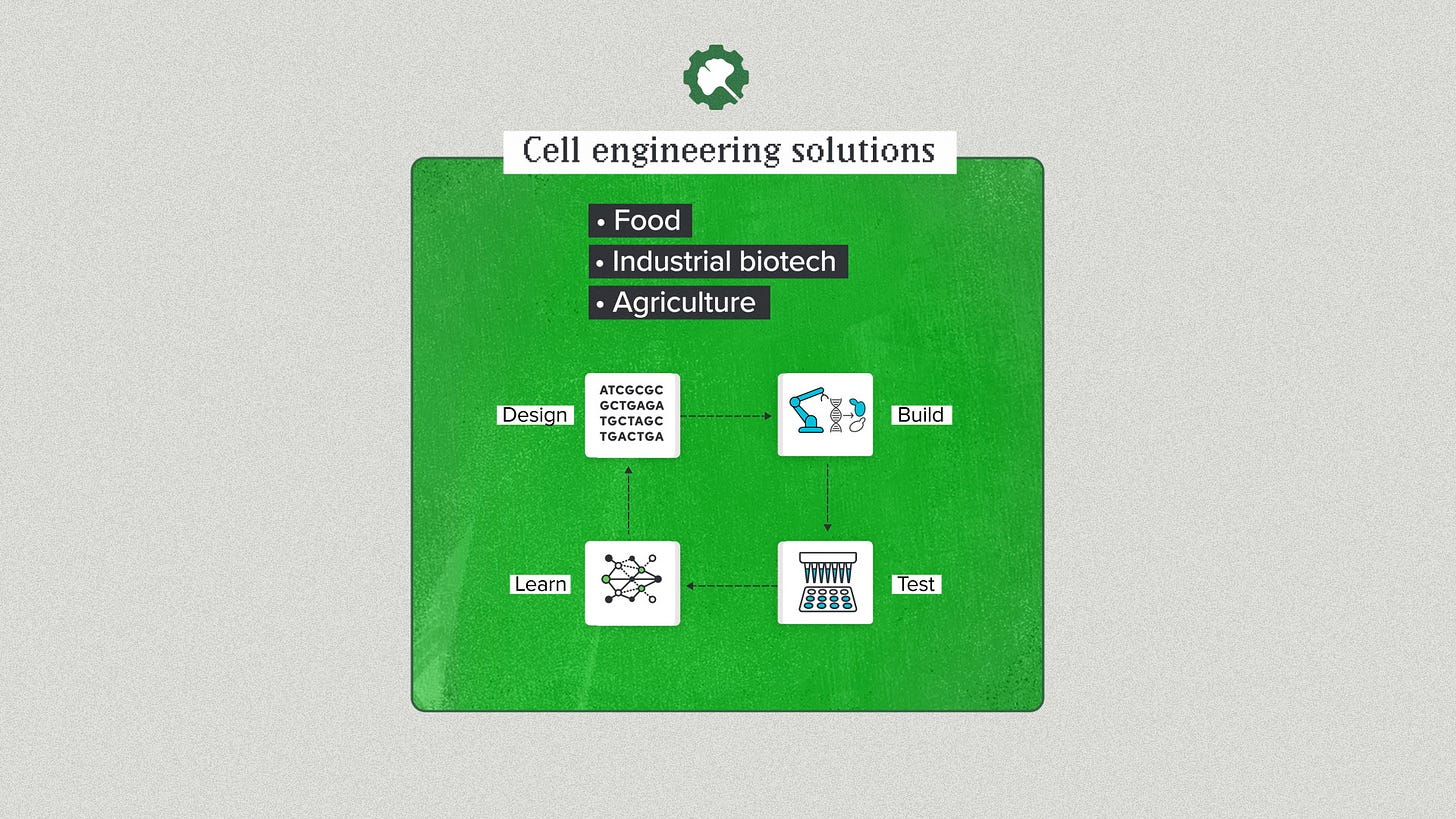
Then, of course, there's the whole enchilada. Design, Build, Test & Learn. You can come to Ginkgo for a complete cell engineering solution. You need yeast to produce a food protein? You need a fungal strain to manufacture an industrial enzyme? You need a soil microbe to fix nitrogen for agriculture? We can handle the entire synthetic biology arm of your R&D project and deliver a strain.
The DBTL cycle isn't the only way to subdivide biotech R&D. I've seen project teams organized by expertise: bioinformatics, biochemistry, pharmacology, etc. I've seen R&D organized in terms of linear progress: the technology readiness levels, the stages of drug development. We'll probably need multiple frameworks to cover the complexity of biology.
But for me, the lesson of DBTL is that what makes these things work well operationally is an emphasis on the interface. We don't want to organize the processes by how they work internally, we organize by how they connect to each other.
Design outputs a digital DNA sequence file. Build outputs engineered DNA. Ginkgo Datapoints outputs a collection of experimental data. Ginkgo Solutions outputs an engineered microbe. These are discrete, well-defined, exchangeable objects and data types. Our foundry users, our customers, can integrate these services into their local R&D programs and build with the outputs.